Sayne Park1, Yoon Ho Choi2*
1 University of Wisconsin-Madison, 333 East Campus Mall #11101, Madison, WI 53715-1384
2 Dept of AI and Informatics, Mayo Clinic, Griffin, 4500 San Pablo Rd S, Jacksonville, FL 32224
* yh.choi.phd@gmail.comAbstract
The expansion into 3D visualization has played a crucial role in biological and medical research. Researchers use this technology to understand complex cellular dynamics more effectively. Blender, Unity and Unreal Engine have become leading tools for 3D cell visualization. Each platform offers distinct capabilities and strengths that address different visualization needs. This paper examines the functionality of these three platforms by analyzing specific examples. By focusing on their technological features and unique advantages, the paper provides a practical guide on how each platform is utilized in biological research scenarios. This guide might help researchers select the most appropriate platform for their 3D cell visualization projects.
Author Summary
We explored how three popular tools—Blender, Unity and Unreal Engine—can help scientists create detailed 3D visuals of cells and biological processes. Our study highlights how Blender excels in making highly accurate static models, Unity supports real-time simulations and Unreal Engine provides stunning visual quality for large-scale data. By comparing these platforms, we aim to guide researchers in choosing the right tool for their projects. This work emphasizes how 3D visualization can make complex biological concepts more accessible, improving both research and education.
Introduction
Three-dimensional (3D) visualization has become an essential tool in biological and medical research, significantly contributing to the advancement of understanding of complex cellular dynamics (Borrel and Fourches 2017; Kim et al. 2023; Martinez et al. 2021; Kozlíková et al. 2017). Researchers rely on this technology to create accurate, interactive and immersive representations of 3D cell behaviors (Borrel and Fourches 2017; Martinez et al. 2021). These representations help overcome practical limitations in research and education contexts (Franzluebbers et al. 2022; Kim et al. 2023; Kozlíková et al. 2017).
Blender, Unity and Unreal Engine represent three leading platforms for 3D cell visualization (Borrel and Fourches 2017; Chen 2023; Zhang et al. 2019). Blender has traditionally been the preferred choice for static and semi-dynamic visualizations (Andrei et al. 2011; Garwood and Dunlop 2014), as researchers frequently use it to create detailed and structurally accurate models (Andrei et al. 2011; Andrei et al. 2012; Zhang et al. 2019). Blender’s open-source nature, comprehensive toolset and adaptability have established it as a staple in scientific visualization (Andrei et al. 2011; Andrei et al. 2012; Zhang et al. 2019). It is especially effective for generating high-fidelity, static models of cellular structures and molecular processes (Andrei et al. 2011; Garwood and Dunlop 2014).
Conversely, Unity and Unreal Engine, originally designed as game engines, gained recognition in scientific fields due to their advanced rendering capabilities and interactive features. These platforms, which were initially developed to support dynamic, real-time simulations in gaming, evolved to enable immersive virtual reality (VR) environments and interactive exploration of biological data (Borrel and Fourches 2017; Chen 2023; Su et al. 2020). The scientific community’s adoption of game engines as modeling software reflects the growing demand for tools that combine static modeling with dynamic, real-time interaction for live-cell visualizations (Borrel and Fourches 2017; Chen 2023).
This paper examines the unique strengths of Blender, Unity and Unreal Engine, in 3D modeling applications (Andrei et al. 2011; Borrel and Fourches 2017; Chen 2023; Su et al. 2020; Garwood and Dunlop 2014). Blender remains the most widely used tool for static modeling (Andrei et al. 2011), while Unity and Unreal Engine excel in dynamic and immersive visualization (Andrei et al. 2011; Chen 2023; Marsden and Shankar 2020; Su et al. 2020). This paper aims to assist researchers in selecting the most appropriate software for their specific visualization needs (Borrel and Fourches 2017; Franzluebbers et al. 2022; Martinez et al. 2021).
Applicable Platforms for the Biological Data Visualization
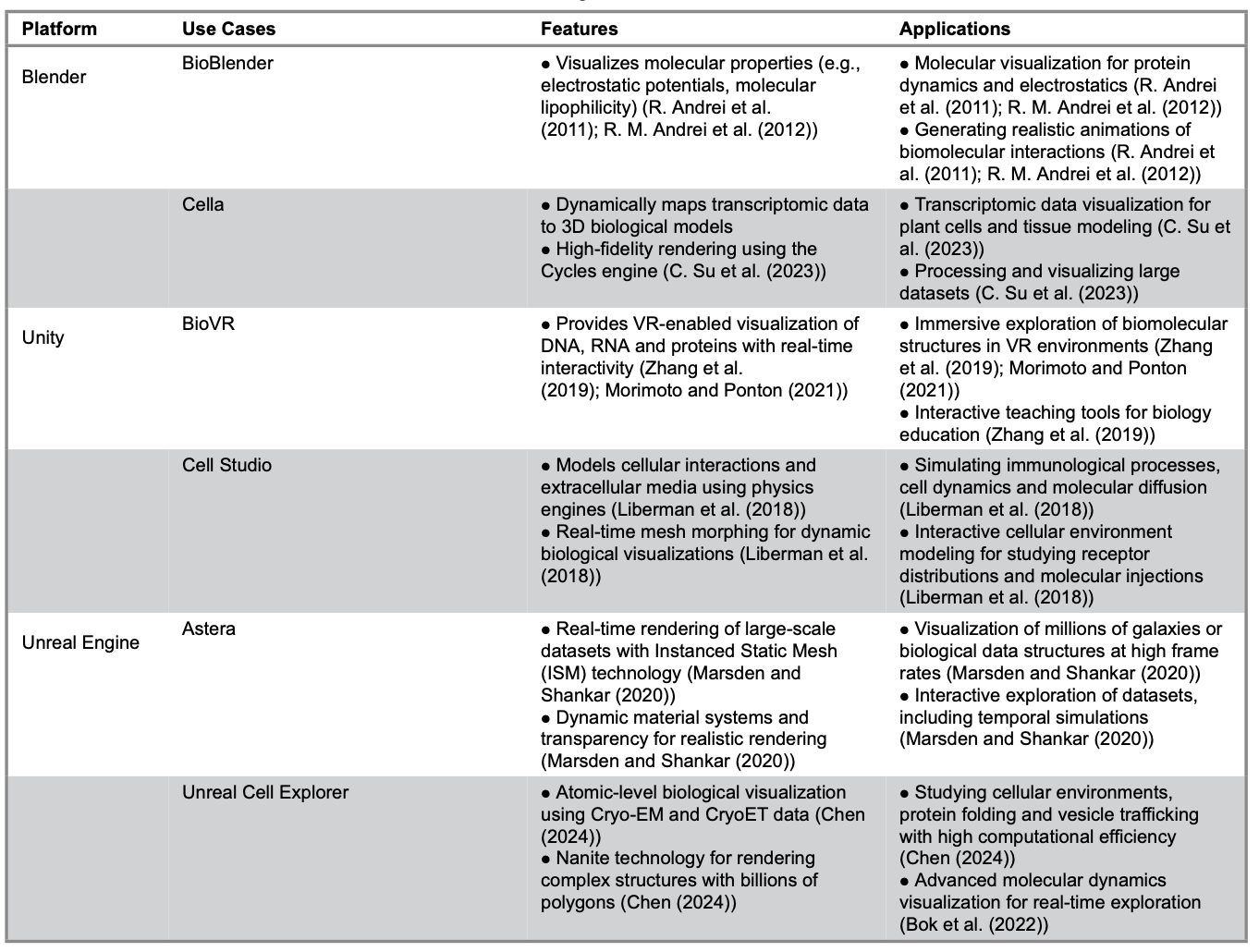
This paper focuses on three platforms applicable to the visualization of biological data: Blender, Unity and Unreal Engine. Table 1 summarizes key information about these platforms, including their use cases, features and potential applications.
Table 1. Platforms and Extensions for 3D Visualization in Biological Research
Blender
Blender serves as a cornerstone platform for biological visualization. It is widely recognized for its open-source flexibility and precision in static and semi-dynamic modeling. Researchers benefit from its powerful Python API, which supports the integration of external datasets (PDB, CSV files, microscopy) seamlessly. This capability allows the creation of high-fidelity representations of biological systems.
Applications like BioBlender showcase Blender's capability to integrate biophysical data with advanced rendering techniques. This integration allows researchers to accurately visualize molecular properties, such as electrostatic potentials. In addition, Blender’s comprehensive toolset offers a variety of accessibility and customization options to suit diverse research needs.
Researchers seeking detailed and customizable visualization solutions consider Blender an essential resource in biological studies.
BioBlender
Blender serves as the foundational platform for BioBlender. It provides robust tools for constructing intricate 3D models and animations of cellular environments, proteins and molecular structures (Andrei et al. 2011; Andrei et al. 2012). Researchers use Blender to facilitate dynamic and interactive visualizations of molecular processes by integrating biophysical and biochemical data. This integration enhances the realism of visualized molecular phenomena.
The Blender Game Engine plays a crucial role in interpolating protein motions while maintaining biophysical constraints (Andrei et al. 2011; Andrei et al. 2012). This functionality enables the generation of realistic transitional forms between experimentally derived models.
Blender’s rendering capabilities are also customized to represent molecular properties, such as molecular lipophilicity and electrostatic potentials. Instead of relying on conventional color maps, researchers use surface textures, such as shiny or rough finishes, to convey hydrophobicity and hydrophilicity effectively.
Blender supports seamless integration with external scientific tools like the adaptive Poisson-Boltzmann Solver (Andrei et al. 2012). These integrations allow researchers to import and visualize data, such as electrostatic fields and hydropathy maps, directly within Blender’s 3D environment. Its advanced rendering features, such as bump mapping and particle animations, illustrate molecular dynamics, electrostatic field lines and intermolecular physicochemical interactions such as electrostatic, hydrophobic and biomolecular binding interactions (Andrei et al. 2011; Andrei et al. 2012). These features produce photorealistic representations that enhance the clarity and accessibility of molecular phenomena.
BioBlender also includes custom tools and interfaces explicitly designed for biologists to simplify manipulating molecular data and ensure the biological accuracy of animations (Andrei et al. 2012). This comprehensive suite of features underscores Blender’s versatility as a platform that bridges computational biology and interactive scientific visualization through media rendering, 3D modeling and animated representation. Blender facilitates a deeper understanding of complex biological phenomena by making molecular interactions visually intuitive and dynamically navigable. This approach enhances accessibility for researchers and educators, broadening its utility across diverse audiences.
Cella
Cella is a Blender-based visualization framework developed for three-dimensional rendering of single-cell transcriptomics data in plant tissues (Su et al. 2023). Researchers extensively utilize Blender within the Cella platform to integrate single-cell transcriptomics data with 3D biological models. Using Blender’s Python scripting module, transcriptomic data are dynamically mapped onto spatial representations of cells, enabling the seamless integration of biological data with visual models.
The Cycles render engine, known for its high-quality image rendering capabilities, renders within times ranging from 1 to 10 seconds per image (Su et al. 2023). This efficiency is crucial for managing large datasets. Additionally, researchers employ Blender to convert two-dimensional (2D) cross-sectional images of biological structures into 3D models by processing STL file formats. This process ensures accurate spatial representation of tissues.
Blender’s Python scripting capabilities enable dynamic adjustments of object colors based on transcriptomic data (Su et al. 2023). These adjustments produce transparent and interpretable visualizations. Researchers also employ the Cell Fracture add-on to enhance the visual realism of cellular geometries (Su et al. 2023), allowing the subdivision of structures and the introduction of natural randomness, reflecting biological complexity.
Researchers improve computational efficiency using Blender to simplify the geometries of 3D models (Su et al. 2023). This approach reduces processing demands while maintaining visual accuracy. In parallel with geometric simplification, the software averages RNA count data within biologically defined clusters and maps these averaged values to corresponding cell collections in Blender. By reducing the dimensionality of single-cell transcriptomic data at the cluster level, this aggregation strategy significantly decreases the computational load required for real-time visualization while preserving biologically meaningful expression patterns. Together, geometric simplification and RNA data averaging streamline the overall visualization pipeline.
Using the Cycles render engine, 42,978 images were generated with optimized settings to ensure consistent output across the dataset (Su et al. 2023). This workflow demonstrates Blender’s scalability in effectively managing complex, high-resolution biological datasets, establishing it as a powerful tool for large-scale studies in biology.
Comparative Analysis Across Blender’s Use Cases
Blender is a versatile and robust platform for researchers, particularly in biological and scientific fields, offering advanced tools for 3D visualization. It supports detailed 3D modeling and rendering capabilities through tools like BioBlender, making it practical for visualizing intricate molecular structures with their chemical and physical properties (Andrei et al. 2012). Python scripting capabilities further enhance Blender's adaptability, allowing seamless integration of biological datasets, such as transcriptomic data, for dynamic and interpretable modeling (Su et al. 2023).
Jorstad and colleagues highlight how NeuroMorph tools demonstrate Blender's utility in transforming 2D datasets into spatially accurate 3D reconstructions (Jorstad et al. 2018). NeuroMorph facilitates the analysis and visualization of complex axonal structures in 3D, using Blender to reconstruct cellular models from segmented 2D electron microscopy images. These tools enable the measurement of morphometric features, such as bouton volume and synaptic density, within Blender's 3D modeling environment (Jorstad et al. 2018).
Similarly, Kozlíková et al. (2017) highlights the critical role of stereoscopic and dynamic visualization techniques in conveying molecular dynamics effectively, resonating with Blender’s ability to produce scalable and interactive visual models (Kozlíková et al. 2017).
Blender achieves computational efficiency by simplifying 3D geometries while preserving visual fidelity. This strength is evident in the BioBlender project, which uses Blender for molecular visualization. It simulates surface properties and molecular interactions to enhance understanding. Furthermore, the BlendMol plugin, developed by Durrant (Durrant 2019), integrates macromolecular analysis tools like VMD and PyMOL with Blender, an open-source platform widely used for scientific visualization. BlendMol enables seamless import of molecular visualizations and coloring schemes from VMD and PyMOL into Blender, simplifying workflows. By leveraging Blender’s advanced rendering capabilities, BlendMol produces high-quality visuals suitable for publications, outreach programs and educational use (Durrant 2019).
These studies underscore Blender's growing recognition as a leading tool in scientific visualization, bridging technical rigor and aesthetic clarity. Its integration with external platforms further reinforces its role as a central tool in modern molecular and structural biology research.
This comprehensive alignment of Blender's functionalities with the reviewed studies validates its application as a transformative tool in 3D visualization and analysis across multiple disciplines.
Unity
Unity has emerged as a leading platform for biological simulations. Its advanced scripting environment, real-time rendering capabilities and support for immersive technologies set it apart. The following sections examine notable Unity-based applications and their technical implementations, highlighting their transformative role in biological data visualization.
BioVR Platform
The BioVR application uses Unity3D as its foundational platform due to Unity’s comprehensive support for VR development and extensive online resources (Morimoto and Ponton 2021; Zhang et al. 2019). These features make it an accessible and efficient choice for building interactive applications (Zhang et al. 2019). Unity provides essential tools for creating VR-enabled desktop applications with its robust GameObject system serving as the structural backbone for all elements in the application. In BioVR, GameObjects represent biological data, including DNA, RNA and protein structures. Components such as the MeshFilter define geometry, while the MeshRenderer handles the rendering of visual elements (Zhang et al. 2019).
BioVR leverages Unity’s UV mapping and texture management capabilities to display nucleotide sequences dynamically (Zhang et al. 2019). The Texture2D.SetPixel() API assigns colors corresponding to specific nucleotides, rendering them on a scrolling plane geometry. This functionality allows users to explore sequences containing up to 100,000 base pairs (Zhang et al. 2019). These tools enable interactive visualization of complex biological data (Zhang et al. 2019).
The MonoBehaviour class in Unity underpins BioVR’s game loop architecture, managing lifecycle events such as initialization, updates and user interactions (Zhang et al. 2019). Core functions like Awake(), Start() and Update() ensure real-time responses to inputs from the Oculus Rift and Leap Motion devices (Zhang et al. 2019). Unity’s integration with the Oculus SDK enables stereoscopic rendering, which mimics human binocular vision using two offset camera objects. The Leap Motion SDK further extends input capabilities by enabling hand-tracking, providing users with intuitive interaction with biological data (Zhang et al. 2019).
The application adopts a Model-View-Controller (MVC) architecture, inherited from UnityMol, to achieve a clear separation between data management, rendering and user input processing (Zhang et al. 2019). Unity serves as the framework for implementing this architecture. Models represent biological data, views manage 3D visualizations and controllers process user inputs. In addition, the Hover UI Kit, an open-source Unity plugin developed and distributed by the Aesthetic Interactive community via GitHub, creates dynamic menus anchored to the user’s hand, enhancing interactivity in the VR environment (Zhang et al. 2019).
Unity’s physics and shader systems enable dynamic interactions within BioVR (Zhang et al. 2019). For example, the system highlights specific residues on protein structures when users hover their hands over corresponding nucleotide sequences (Zhang et al. 2019). These interactions, supported by Unity’s real-time rendering and physics capabilities, create an
immersive and intuitive user experience. By extending UnityMol’s protein visualization features and integrating VR-specific functionalities, Unity serves as the core platform for BioVR (Zhang et al. 2019). It enables the aggregation and interactive visualization of DNA, RNA and protein structures within a unified 3D environment.
Cell Studio
In the development of “Cell Studio,” Unity3D was chosen as the game engine due to its robust toolset, which includes graphical rendering, scripting support through C# and physics-based capabilities (Liberman et al. 2018). Unity’s cross-platform compatibility enabled the simulation to be compiled and deployed across various computer platforms. This feature enhanced accessibility for users without requiring specialized hardware (Liberman et al. 2018). The platform’s graphical user interface (GUI) was constructed using Unity3D’s GUI tools (Liberman et al. 2018). These tools simulate a 2D canvas that overlays the 3D simulation environment, facilitating intuitive user interaction. Freely available 3D cell models with a mesh structure and graphical texture were incorporated to enhance the platform’s real-time visualization capabilities. The developers used the MegaFiers Unity package to animate the cell models dynamically, enabling real-time morphing of the meshes based on simulation constraints and conditions (Liberman et al. 2018).
Unity’s physics engine played a critical role in modeling spatial and physics-based phenomena (Liberman et al. 2018). Researchers used the engine to simulate the extracellular medium and manage cell movements within the 3D environment, offering a realistic and dynamic framework for simulating interactions between cells and their environment. Visualization of real-time data, such as cell counts and receptor distributions, was achieved using the Graph Maker Unity package (Liberman et al. 2018). This allowed users to monitor simulation outcomes through dynamic graph plotting.
The platform also provided precise user control over various aspects of the simulation. The timeline could be manipulated through actions such as playing, pausing, rewinding and fast-forwarding, and molecules could be injected into the extracellular medium in real time. These interactive features were enabled through Unity’s integrated scripting capabilities, ensuring seamless integration of user commands and simulation responses. The data frames produced by the simulation’s backend server were parsed and rendered into real-time graphical simulations that were visually coherent and biologically relevant.
Unity’s inherent cross-platform support ensured that the platform could be deployed on various systems, increasing its accessibility to researchers and reducing technical barriers to its adoption. Through these features, Unity3D served as the backbone of "Cell Studio" (Liberman et al. 2018) enabled the creation of an interactive, scalable and visually rich simulation platform for studying immunological processes.
Comparative Analysis across Unity’s Use Cases
Unity3D is a comprehensive and versatile platform for researchers and developers, particularly in creating interactive and visually engaging biological simulations (Liberman et al. 2019). Its advanced graphical rendering engine and robust physics engine make it a prime choice for accurately representing complex biological systems, including molecular structures, cellular dynamics and extracellular environments. This capability is reinforced by Unity's support for scripting in C#, enabling real-time interactions and seamless parameter adjustments during simulations (Liberman et al. 2018).
Researchers highlight Unity’s flexibility in studies such as Eriksen et al. (2020), who explore augmented reality (AR) applications that integrate Unity for creating interactive 3D molecular models, emphasizing its role in fostering user engagement and educational advancements. Similarly, Gandhi and colleagues (2020) discuss the use of Unity for real-time interactive simulations of organic molecules, highlighting the platform’s capacity for dynamic visualization and manipulation of molecular structures, which enhances the comprehensibility of complex biological processes (Gandhi et al. 2020).
Unity’s integration with third-party tools and plugins further extends its capabilities. VR functionalities, enabled through the Oculus SDK and Leap Motion support hand-tracking features (Zhang et al. 2019), align with findings from the study on UnityMol (Lv et al. 2013). This study demonstrates Unity's effectiveness in creating immersive and interactive experiences for exploring molecular flexibility (Lv et al. 2013). Features like dynamic mesh morphing were achievable with plugins such as MegaFiers, allowing for real-time animation of biological phenomena, supporting accurate and realistic depictions (Liberman et al. 2018). This aligns with studies showcasing Unity’s application in dynamically visualizing cellular mechanics and molecular assemblies.
Unity’s cross-platform compatibility ensures wide accessibility and deployment, facilitating scalability for large datasets and complex biological systems. Liberman et al. (2019) emphasizes Unity’s strength in modeling cell-surface interactions, utilizing its GUI tools to develop intuitive interfaces for real-time data visualization and parameter adjustments (Liberman et al. 2019).
These capabilities make Unity a scalable and user-friendly platform for tackling computationally intensive tasks. Various researchers echo this sentiment, focusing on the role of Unity in scientific visualization.
Unity’s real-time data processing and rendering capabilities establish it as a transformative tool in biological research. This platform integrates technical complexity with usability and accessibility; Unity facilitates innovative exploration of emergent behaviors in biological and dynamic systems. The related studies highlight Unity’s pivotal role in advancing scientific communication and education through high-quality visualizations and interactive tools (Durrant 2019; Jorstad et al. 2018; Liberman et al. 2018; Liberman et al. 2019; Lv et al. 2013; Morimoto and Ponton 2021; Zhang et al. 2019).
Unreal Engine
Unreal Engine is well-known for its high-fidelity graphics and advanced rendering capabilities. Researchers utilize this platform to develop scientific visualization projects that require exceptional visual quality and computational depth. Its real-time rendering, combined with robust physics simulation and extensibility, facilitates the creation of immersive and interactive scientific applications.
The following sections explore key use cases for Unreal Engine. These examples illustrate its technical features and transformative impact on scientific visualization.
Astera
Marsden and Shankar (2020) select Unreal Engine 4 (UE4) as the foundation for this study due to its advanced suite of tools. These tools include real-time rendering, animation, physics-based modeling, artificial intelligence, networking and parallel processing capabilities, which make it suitable for high-performance visualizations. The simulation utilized Unreal Engine’s Instanced Static Mesh (ISM) technology to optimize GPU performance by combining identical objects into a single draw call. This approach enables the visualization of millions of galaxies in a cosmological volume while maintaining high frame rates, reaching up to 60 frames per second (FPS) on advanced GPUs (Marsden and Shankar 2020).
Galaxy models in the simulation were represented using static meshes to balance computational efficiency with visual fidelity (Marsden and Shankar 2020). Spiral galaxies were modeled as simple two-polygon planes, while elliptical galaxies were designed as diffuse spheroids (Marsden and Shankar 2020). Unreal Engine’s material system enhanced realism by incorporating transparency and dynamic textures. Galaxy images sourced from the Sloan Digital Sky Survey (SDSS) were converted into textures and applied as materials, ensuring visually accurate representations (Marsden and Shankar 2020). The material system also supported dynamic simulations, such as Active Galactic Nuclei (AGN) activity, which visualized using oscillating brightness functions to mimic time-variable behavior (Marsden and Shankar 2020).
The rendering engine in Unreal Engine processed and displayed large-scale datasets efficiently. Galaxies were assigned properties such as TType (morphological classification) and stellar mass using C++ scripts, which dynamically spawned galaxies and matched them to simulation data (Marsden and Shankar 2020). Lighting and material effects further enhanced realism, while Unreal Engine’s alpha channels created smooth transitions and eliminated sharp edges, providing a visually seamless experience (Marsden and Shankar 2020).
Unreal Engine’s real-time rendering capabilities allowed interactive exploration of the cosmic web and users could zoom, pan and rotate within the 3D simulation using camera controls (Marsden and Shankar 2020). Preliminary VR support was integrated through Unreal Engine’s VR tools, enabling immersive binocular 3D visualization of the universe.
The platform’s performance optimization relied on Unreal Engine’s GPU-driven architecture (Marsden and Shankar 2020). This architecture minimized CPU draw calls by using batch processing and instancing techniques (Marsden and Shankar 2020). These optimizations ensured scalability, allowing the simulation to run effectively across various hardware configurations and support different galaxy catalog sizes while maintaining smooth operation.
Unreal Engine’s flexibility and powerful toolset provided opportunities for future development. These opportunities include the addition of dark matter visualization, time-evolution simulations and gravitational lensing effects. These features demonstrate Unreal Engine’s suitability for large-scale scientific visualization projects, particularly for applications requiring real-time rendering and interactive exploration.
Unreal Cell Explorer
Unreal Engine demonstrates significant potential for biological visualization, particularly through its real-time rendering and dynamic material systems. To exemplify this, we describe its use for cellular visualization under the conceptual name ‘Unreal Cell Explorer,’ which encompasses its use in integrating Cryo-EM data, modeling lipid bilayers and rendering atomic-level interactions. Unreal Engine 5 (UE5) further enhances these capabilities by leveraging its virtualized geometry system, Nanite, which dynamically adjusts the level of detail based on the viewer’s perspective. Researchers use Nanite to enable real-time rendering of billions of triangles while maintaining computational efficiency, supporting the visualization of complex biological systems, such as atomic-level macromolecular environments, on consumer-grade hardware like Nvidia RTX 4070 GPUs (Chen 2023).
UE5 supports interactive exploration of cellular environments. Users can seamlessly navigate through keyboard, mouse or controller inputs, which are further enhanced by user-defined interaction protocols (Chen 2023). These protocols include features such as movement adjustments and pass-through-wall functionalities.
UE5 facilitates the integration of Cryo-EM and CryoET datasets through customized Python scripts. These scripts enable the precise placement of macromolecules based on their experimental coordinates, orientations and conformations (Chen 2023). Protein structures sourced from the Protein Data Bank are imported into UE5 via UCSF ChimeraX (Chen 2023). Researchers convert these structures into compatible mesh formats and optimize them using Nanite (Chen 2023). Cellular membranes and organelles are modeled parametrically as lipid bilayers to ensure realistic structural representation (Chen 2023). Additionally, UE5 supports dynamic real-time annotation. In-game tools identify molecular features and display relevant metadata (Chen 2023).
UE5’s blueprint system allows the animation of molecular dynamics, such as protein conformational changes or filament movements (Chen 2023). Researchers use this system to control motion paths and interaction behaviors precisely. The system also facilitates rendering symmetric protein complexes and helical structures through automated duplication and spatial alignment (Chen 2023). The rendering engine incorporates advanced material systems to enhance visual fidelity, supporting transparency, dynamic textures and interaction-based modifications.
UE5’s scalability and cross-platform compatibility make it suitable for various applications, ranging from large-scale scientific visualizations to educational tools. Ongoing efforts to integrate VR support aim to enhance user immersion, further expanding the platform’s potential as a framework for high-performance biological simulations and interactive exploration. These attributes underline Unreal Engine’s versatility and power in providing innovative solutions for complex visualization challenges in the life sciences.
Comparative Analysis across Unreal Engine’s Use Cases
Unreal Engine is a robust and versatile platform researchers widely use to create interactive and high-fidelity simulations. Its advanced rendering engine, real-time global illumination system and physics-based modeling capabilities make it a prime choice for accurately representing intricate systems, including molecular structures and biological dynamics. Unreal Engine’s Blueprint Visual Scripting and Python API enhance these strengths, enabling seamless integration and dynamic parameter adjustments during simulations (Chen 2023; Marsden and Shankar 2020).
The adaptability of Unreal Engine is exemplified in studies such as Trenchev et al. (2024), where Unreal Engine was used in conjunction with MATLAB and NAMD to facilitate real-time genomic data visualization and simulation (Trenchev et al. 2024). This integration enables detailed rendering of molecular structures and dynamic exploration of biological processes,
emphasizing the platform’s role in scientific discovery.
Similarly, Bok and colleagues (2022) demonstrated the application of Unreal Engine 4 in visualizing molecular dynamics simulations, utilizing its capabilities for comparative visual analysis and interactive exploration of protein conformations (Bok et al. 2022).
Unreal Engine’s support for advanced visualization technologies extends its potential in integrating molecular and genomic data. The platform’s real-time rendering capabilities and adaptability to integrate external tools, like Python and NAMD, enable efficient visualization of intricate biological systems, including molecular assemblies and cellular structures. This integration emphasizes Unreal Engine's scalability and effectiveness in rendering complex datasets and facilitating scientific discovery (Trenchev et al. 2024).
Unreal Engine’s real-time rendering, scalability and integration capabilities position it as a transformative tool for advancing biological and molecular research. By bridging complex datasets with user-friendly interactivity, Unreal Engine enables innovative explorations of biological phenomena and fosters scientific communication and education advancements. These studies underscore its pivotal role in delivering high-quality visualizations and interactive tools for researchers and educators alike.
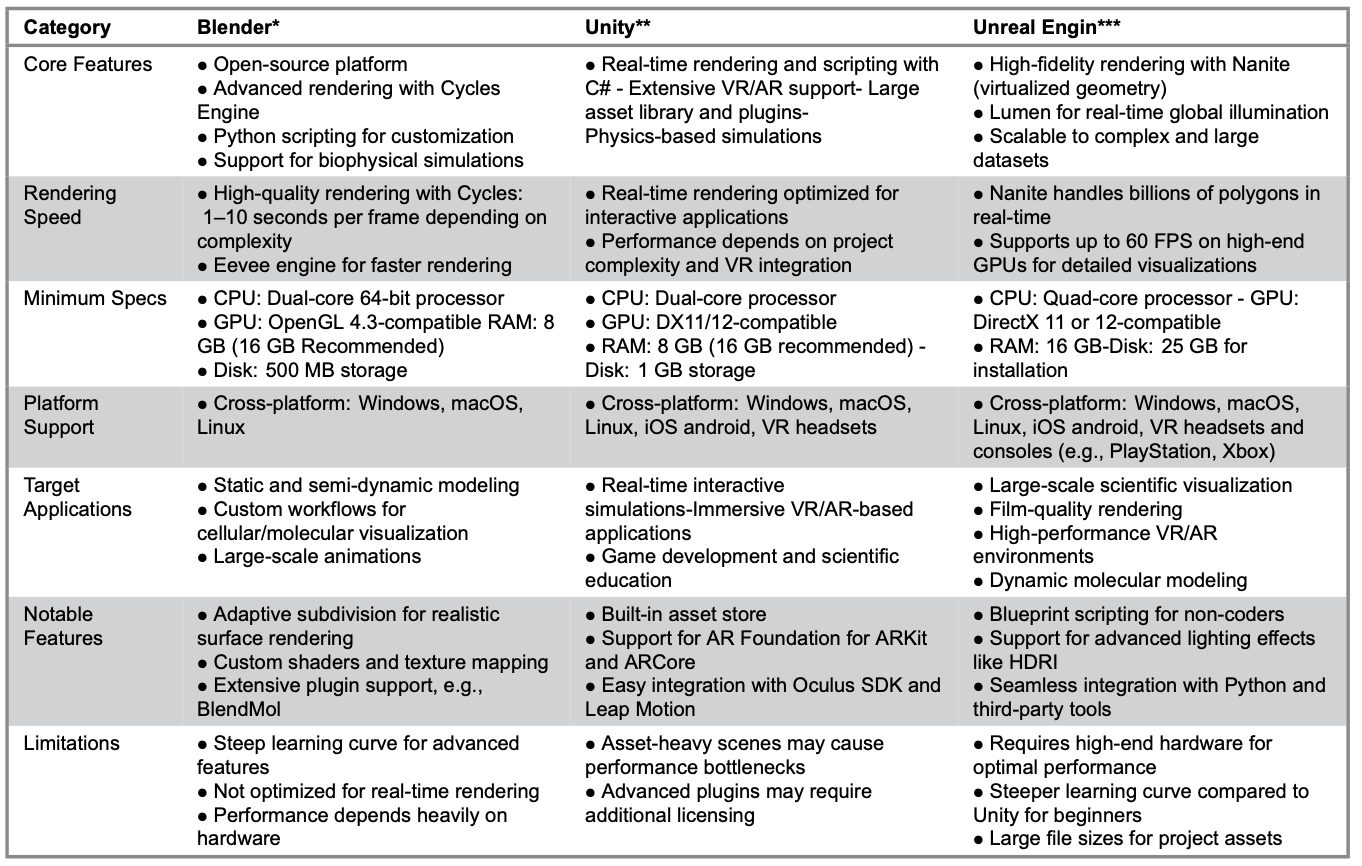
Table 2 provides a detailed comparison of Blender, Unity and Unreal Engine, focusing on their core features, rendering capabilities, system requirements, platform support, target applications, notable features and limitations.
Table 2. Blender, Unity and Unreal Engine: Technical Features and Specifications
Note. References marked with * are from the following official websites: *Blender Official Website. Available from: https://www.blender.org/ *Unity Official Website. Available from: https://unity.com/ **Unreal Engine Official Website. Available from: https://www.unrealengine.com/
Contributions and Insights of 3D Visualization Modeling Tools
Blender, Unity and Unreal Engine transform live-cell imaging by enabling precise, interactive and temporally dynamic visualizations. Blender excels in modeling high-fidelity cellular structures and integrating complex biological datasets. For example, tools like Cella can map transcriptomic data onto spatially accurate 3D geometries (Su et al. 2023). These capabilities advance research on tumor heterogeneity and organ development, although real-time applications remain a challenge due to computational limitations (Su et al. 2023).
Unity and Unreal Engine extend these capabilities by adding interactivity and real-time simulation to live-cell imaging. Unity’s VR-enabled platforms, such as BioVR, provide immersive exploration of biomolecular structures, while Unreal Engine’s high-fidelity rendering and dynamic simulations support detailed studies of cellular behaviors like protein folding and vesicle trafficking (Chen 2023; Zhang et al. 2019). Temporal visualization, or 4D imaging, has become essential for capturing dynamic cellular processes such as mitosis and intracellular signaling, bridging theoretical models and experimental data (Lv et al. 2013; Trenchev et al. 2024).
These platforms have also revolutionized traditional visualization methods. Blender’s precision allows for creating high-quality static and semi-dynamic models, essential for analyzing molecular conformations and cellular geometry (Andrei et al. 2012; Su et al. 2013). Unity and Unreal Engine enable researchers to create immersive environments for collaborative exploration through their VR and real-time rendering capabilities, making these tools invaluable for educational applications and interdisciplinary studies (Chen 2023; Gandhi et al. 2020).
Future advancements in artificial intelligence (AI), augmented reality (AR) and hardware technologies are poised to significantly enhance 3D visualization platform capabilities. AI-driven tools are enhancing workflows by automating tasks such as annotation and segmentation. OpenAnnotate3D exemplifies this trend, enabling automatic generation of 2D and 3D annotations for multi-modal data. While significantly improving annotation efficiency, manual input may still be required for refinement (Zhou et al. 2024). Additionally, studies have explored the use of generative AI for visualization, indicating that AI can assist in creating and refining complex visual data representations (Ye et al. 2024).
In the realm of AR, integrating virtual models with real-world experimental environments opens new avenues for hybrid research. Su et al. (2020) demonstrated that AR applications can support data analysis and assessment in scientific and engineering fields, enhancing interactivity and improving the teaching and learning process. Moreover, the development of reference architectures for AR integration in remote experimentation environments has the potential to verify the effectiveness of immersive technologies in educational settings (Nardi da Silva et al. 2022).
Recent advancements in hardware technologies, particularly in GPUs and VR headsets, markedly enhanced the capabilities and accessibility of advanced visualization platforms (Marsden and Shankar 2020). Modern GPUs now possess significantly improved real-time rendering capabilities, enabling complex simulations and interactive visualizations. Agarwal et al. (2024) highlight these advancements in their ability to enhance computational efficiency, support high-fidelity rendering and widely apply in scientific and computational research (Agarwal et al. 2024).
In the context of education, VR integration has demonstrated transformative potential by providing immersive and interactive learning experiences. Recent research underscores the value of VR in simulating real-world scenarios, fostering active learning and enhancing knowledge retention through immersive engagement (Gandhi et al. 2020). These advancements facilitate the development of novel pedagogical methodologies, allowing students to explore complex concepts through experiential learning, which traditional approaches often struggle to achieve (Vats and Joshi 2023).
The continuous evolution of GPU architectures and VR technologies has further mitigated computational barriers, broadening their applicability across a broader range of disciplines.
These advancements, supported by empirical studies (Marsden and Shankar 2020; Lv et al. 2013), show how enhanced hardware performance enables sophisticated visualizations that were previously computationally prohibitive. These improvements ensure higher-quality simulations and visual outputs and foster the development of interdisciplinary applications, expanding the reach and utility of advanced visualization platforms in research, education and professional domains.
Collectively, these advancements in AI, AR and hardware technologies are expected to drive innovation in 3D visualization platforms, enabling more efficient workflows, immersive experiences and accessible tools for researchers and educators alike. While challenges remain, including high computational demands and complex workflows, ongoing efforts to optimize performance and simplify interfaces will broaden the accessibility of these tools. By addressing these limitations, Blender, Unity and Unreal Engine will continue to drive innovation in cellular research and education, enabling deeper insights into live-cell dynamics and complex biological processes. These advancements ensure that 3D and live-cell visualization technologies remain at the forefront of innovation, shaping the future of biological research and medical applications.
Conclusions
The development of 3D and live-cell visualization technologies has introduced transformative opportunities in cellular and molecular research, with different platforms offering specialized strengths suited to distinct visualization needs. The characteristics of Blender, Unity and Unreal Engine, grounded in their underlying technologies, define their suitability for specific 3D and live-cell visualizations.
Blender, optimized for high-fidelity static and semi-dynamic visualizations, is ideal for creating detailed 3D cellular models and molecular structures. This platform excels in precision rendering, allowing researchers to integrate complex biological datasets such as transcriptomics or molecular interactions into visually accurate and interpretable models. Blender is best suited for projects focusing on static representations of cellular geometries or molecular dynamics, such as analyzing molecular conformations or exploring structural variations across populations of cells.
With its advanced real-time rendering and scripting capabilities, Unity provides the interactivity necessary for exploring dynamic cellular processes. This platform enables researchers to simulate and visualize live-cell events, such as vesicle trafficking, protein folding or signaling cascades, in real-time. Unity’s ability to incorporate immersive technologies such as VR and AR makes it particularly valuable for real-time educational applications and collaborative exploration of cellular behaviors.
Unreal Engine, designed for high-fidelity visualizations of large-scale datasets and computationally intensive processes, is essential for studying highly detailed and dynamic phenomena. Its tools support the visualization of live-cell processes at multiple scales, from organelle dynamics to atomic-level interactions within macromolecules. Unreal Engine is most effective in applications requiring temporal resolution and spatial detail, such as tracking cellular responses to stimuli over time or simulating intracellular environments under experimental conditions.
By leveraging the distinctive technological strengths of Blender, Unity and Unreal Engine, researchers can address diverse challenges in 3D and live-cell visualization. The alignment of platform characteristics with visualization objectives ensures that these tools not only enhance the understanding of cellular dynamics but also facilitate innovative approaches to studying complex biological systems.
References
Agarwal, T., Gupta, P. and BN, K.R. (2024) ‘Investigating recent developments in GPU-based rendering techniques for graphics and visualization’, 2024 3rd International Conference for Advancement in Technology (ICONAT), IEEE, pp. 1-6.
Andrei, R., Pan, M., Zini, M.F., Zoppe, M. (2011) BioBlender: Visualizing Biology Using Blender, (Online), available at: https://www.scivis.it/images/stories/PDFarticle/PosterEMBO2011.pdf
Andrei, R.M., Callieri, M., Zini, M.F., Loni, T., Maraziti, G., Pan, M.C. and Zoppè, M. (2012) ‘Intuitive representation of surface properties of biomolecules using BioBlender’, BMC bioinformatics, 13(Suppl 4), S16.
Blender Official Website (Online), available at: https://www.blender.org/
Bok, M., Schäfer, M., Brich, N., Schreiner, K., Fäßler, V., Keckeisen, M., Kohlbacher, O. and Krone, M. (2022) ‘Comparative visual analysis of molecular dynamics’, in Eurographics workshop on Visual Computing for Biology and Medicine, Vienna, Austria: VCBM.
Borrel, A. and Fourches, D. (2017) ‘RealityConvert: a tool for preparing 3D models of biochemical structures for augmented and virtual reality’, Bioinformatics, 33(23), pp.3816-3818.
Chen, M. (2024) ‘Rendering protein structures inside cells at the atomic level with Unreal Engine’, bioRxiv, 2023-12.
Durrant, J.D. (2019) ‘BlendMol: advanced macromolecular visualization in Blende’, Bioinformatics, 35(13), pp.2323-2325.
Eriksen, K., Nielsen, B.E. and Pittelkow, M. (2020) ‘Visualizing 3D molecular structures using an augmented reality app’, Journal of Chemical Education, 97(5), pp. 1487-1490.
Franzluebbers, A., Li, C., Paterson, A. and Johnsen, K. (2022) ‘Virtual reality point cloud annotation’, In Proceedings of the 2022 ACM Symposium on Spatial User Interaction, pp. 1-11.
Gandhi, H.A., Jakymiw, S., Barrett, R., Mahaseth, H. and White, A.D. (2020), ‘Real-time interactive simulation and visualization of organic molecules’ Journal of Chemical Education, 97(11), pp.4189-4195.
Garwood, R. and Dunlop, J. (2014) ‘The walking dead: Blender as a tool for paleontologists with a case study on extinct arachnids’, Journal of Paleontology, 88(4), pp.735-746. doi:10.1666/13-088
Jorstad, A., Blanc, J. and Knott, G. (2018) ‘NeuroMorph: a software toolset for 3D analysis of neurite morphology and connectivity’, Frontiers in neuroanatomy, 12, p.59.
Kim, K., Yang, H., Lee, J. and Lee, W.G. (2023) ‘Metaverse wearables for immersive digital healthcare: a review’, Advanced Science, 10(31), p.2303234.
Kozlíková, B., Krone, M., Falk, M., Lindow, N., Baaden, M., Baum, D., Viola, I., Parulek, J. and Hege, H.C. (2017) ‘Visualization of biomolecular structures: State of the art revisited’, Computer Graphics Forum, 36(8), pp. 178-204.
Liberman, A., Kario, D., Mussel, M., Brill, J., Buetow, K., Efroni, S. and Nevo, U. (2018) ‘Cell studio: A platform for interactive, 3D graphical simulation of immunological processes’, APL bioengineering, 2(2).
Liberman, A., Mussel, M., Kario, D., Sprinzak, D. and Nevo, U. (2019) ‘Modelling cell surface dynamics and cell–cell interactions using Cell Studio: A three-dimensional visualization tool based on gaming technology’, Journal of the Royal Society Interface, 16(160), p.20190264.
Lv, Z., Tek, A., Da Silva, F., Empereur-Mot, C., Chavent, M. and Baaden, M. (2013) ‘Game on, science-how video game technology may help biologists tackle visualization challenges’, PloS one, 8(3), p.e57990.
Marsden, C. and Shankar, F. (2020) ‘Using unreal engine to visualize a cosmological volume’, Universe, 6(10), p.168.
Martinez, X. and Baaden, M. (2021) ‘UnityMol prototype for FAIR sharing of molecular-visualization experiences: from pictures in the cloud to collaborative virtual reality exploration in immersive 3D environments’, Acta Crystallographica Section D: Structural Biology, 77(6), pp.746-754.
Morimoto, J. and Ponton, F. (2021) ‘Virtual reality in biology: could we become virtual naturalists?’, Evolution: Education and Outreach, 14(1), p.7
Nardi da Silva, I., García-Zubía, J. and Hernández-Jayo, U. (2022) ‘Definition of a Reference Architecture for the Integration of Augmented Reality/Virtual Reality Techniques in Remote Experimentation Environments’, In the International Conference on Technological Ecosystems for Enhancing Multiculturality (pp. 1354-1361). Singapore: Springer Nature Singapore.
Su, C., Lyu, M., Mähönen, A.P., Helariutta, Y., De Rybel, B. and Muranen, S. (2023) ‘Cella: 3D data visualization for plant single-cell transcriptomics in Blender’, Physiologia Plantarum, 175(6), p.e14068.
Su, S., Perry, V., Bravo, L., Kase, S., Roy, H., Cox, K. and Dasari, V.R. (2020) ‘Virtual and augmented reality applications to support data analysis and assessment of science and engineering’, Computing in Science & Engineering, 22(3), pp.27-39.
Trenchev, I., Trencheva, T., Angelov, V., Spirov, Y., Karshiyska, Y. and Shumanova, K. (2024) ‘Integrating mixed reality with neural networks for advanced molecular visualization in bioinformatics: A mathematical framework for drug discovery’, In ENVIRONMENT. TECHNOLOGIES. RESOURCES., Proceedings of the International Scientific and Practical Conference, 2, pp. 286-292.
Unity Official Website (Online), available at: https://unity.com/.
Unreal Engine Official Website (Online), available at: https://www.unrealengine.com/.
Vats, S. and Joshi, R. (2023) ‘The impact of virtual reality in education: A comprehensive research study’, In the International Working Conference on Transfer and Diffusion of IT (pp. 126-136). Cham: Springer Nature Switzerland.
Ye, Y., Hao, J., Hou, Y., Wang, Z., Xiao, S., Luo, Y. and Zeng, W. (2024) ‘Generative AI for visualization: State of the art and future directions’, Visual Informatics, 8(2), pp.43-66.
Zhang, J.F., Paciorkowski, A.R., Craig, P.A. and Cui, F. (2019) ‘BioVR: a platform for virtual reality assisted biological data integration and visualization’, BMC bioinformatics, 20(1), pp.78.
Zhou, Y., Cai, L., Cheng, X., Gan, Z., Xue, X. and Ding, W. (2024) ‘Openannotate3d: Open-vocabulary auto-labeling system for multi-modal 3d data’, In 2024 IEEE International Conference on Robotics and Automation (ICRA) (pp. 9086-9092). IEEE.