Brenda Moita and Dr. Vikram Sharma
School of Biomedical Sciences, Faculty of Health: Medicine, Dentistry and Human Sciences, Drake Circus, Plymouth PL4 8AA
2School of Biomedical Sciences, Faculty of Health: Medicine, Dentistry and Human Sciences, DRF122, Derriford Research Facility, Drake Circus, Plymouth, PL4 8AA
ABSTRACT
Prostate cancer is the second most prevalent cancer in men and one of the leading causes of mortality globally. Therefore, clinical prostate cancer therapy requires better prognosis and treatment methods. Research on proteomics has enhanced the understanding of the processes underlying tumorigenesis, cancer cell migration, and metastasis. Considering that proteins are the drivers of most cellular responses and the targets for drug delivery, a methodical analysis of the proteome alterations taking place during the initiation and development of prostate cancer might lead to scientific breakthroughs. This review is a systematic literature search to retrieve primary research articles related to prostate cancer proteomics disease mechanisms and aims to discuss and compare current proteomics methods in both the clinical and research context. Aberrant regulation of lipid metabolism due to abnormal expression of the fatty acid synthase (FASN) enzyme has been extensively documented in available literature. An abnormal increase in the expression of this enzyme was associated with prostate cancer. Lastly, prostate cancer diagnosis and prognosis has been enhanced through the use of biomarkers. The use of prostate cancer biomarkers has also been extended to biofluid analysis of extracellular vesicles. Furthermore, profiling of total proteins in prostate cancer cells will help the drug development process to optimise drug activity while limiting cytotoxicity to non-target cells.
INTRODUCTION
Prostate cancer (PCa) is one of the most common men’s cancers worldwide. According to the Office for National Statistics (2019) cancer registration report, PCa was the most common cancer in men in the UK with over 40,000 cases diagnosed in 2017 alone. The current methods of determining the correct therapy for a patient diagnosed with PCa depend almost entirely on Prostate-Specific Antigen (PSA) levels, histopathological features, diagnostic imaging, and clinical assessment of the severity of the disease (Jadvar et al., 2020). Recent developments in genomic technology have changed our perception of complex molecular, genetic, metabolomic, epigenetic, and transcriptomic remodelling in PCa. Extensive genomic and transcriptomic analysis has been used to investigate driver mutations and expressional changes in prostate cancer (Felici et al., 2012; Wyatt et al., 2014; Iglesias-Gato et al., 2016; Yuan et al., 2016). Nevertheless, the discoveries into the genetic environment of PCa are yet to have a major effect on the clinical care of PCa patients. Furthermore, these methods cannot detect all molecular alterations that affect the tumour outcome (Ylipaa et al., 2015). Methodologies for proteome-wide quantitative profiling have the ability to reveal previously identified molecular linkages between genotype and phenotype and to promote the discovery of reliable prognostic biomarkers or effective drug targets for enhanced PCa treatment (Mann et al., 2013; Iglesias-Gato et al., 2016).
Understanding molecular events in cancer requires thorough investigation of the proteome (Boja and Rodriguez, 2014). In recent decades, mass spectrometric methods have enabled high throughput analysis of clinical patient samples (Schubert et al., 2017). A significant development for global protein quantification was the incorporation of isobaric tags, which made it possible to measure proteins globally through several samples in a single experiment (Pappireddi et al., 2019). Classification of diseases according to their molecular characteristics has helped to manage other types of cancer (e.g., breast cancer), and it is expected that this might be feasible for PCa with the help of novel proteomics approaches. This review is the result of a systematic Google Scholar literature search to retrieve thirty primary research articles related to prostate cancer proteomics disease mechanisms. Research conducted over the past four years has been reviewed, summarised, and critically evaluated. The aim of this literature review is to compare and contrast existing proteomics approaches in both clinical and research settings.
LITERATURE SEARCH AND REVIEW STRATEGY
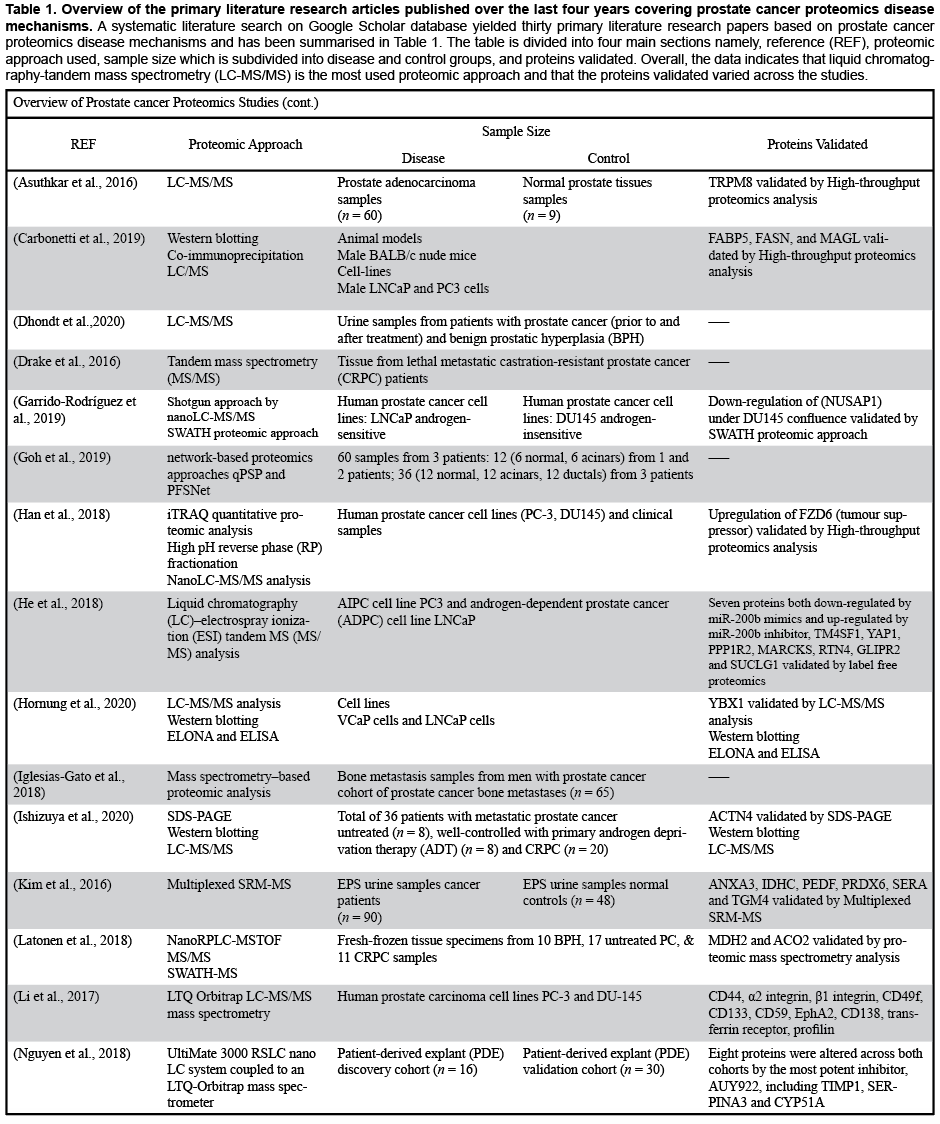
A systematic literature search was conducted via Google Scholar. Records were retrieved based on the following search criteria from Google Scholar database: (1) prostate cancer proteomics disease mechanisms and (2) timespan: 2016-2020. As shown in Table 1, the search resulted in the retrieval of 30 primary literature research articles.
PROSTATE CANCER PROTEOMICS DISEASE MECHANISMS
Prostate cancer (PCa) is a highly heterogeneous disorder; several patients display an aggressive form of the disease with progression and metastasis, whereas other patients display an indolent form of it, having low propensity for progression (Wang et al., 2018). Scientists have identified three stages of human prostate tumours which include: intraepithelial neoplasia, adenocarcinoma androgen-dependent, and adenocarcinoma androgen-independent or castration-resistant. Over the years, developments in molecular technology have made very rapid progress, contributing to the better understanding of the proteomics and genomics events responsible for the initial development and progression of prostate cancer. Studies have shown that the genome of prostate cancer displays a relatively low rate of mutations compared to other cancers (Fiñones et al., 2013; Testa et al., 2019). Unlike genomic and transcriptomic technologies, proteomic technologies based on mass spectrometry (MS) allow comprehensive and direct protein analysis and have therefore been extensively used in the proteomic profiling of clinical specimens (Murray et al., 2017).
Prostate cancer stems from the loss of the regulation of lipid metabolism pathways. Both synthesised and diet acquired lipids play a significant role in the development of prostate tumours. At the proteomic level, enzymes of lipid metabolism have been linked to metastasis of prostate cancer. For instance, proteomic analysis using Western Blot and co-immunoprecipitation Liquid Chromatography–Mass Spectrometry (LC/MS) have heavily implicated the enzyme fatty acid synthase (FASN) in the metastasis of prostate cancer (Carbonetti et al., 2019). Researchers have reported a remarkable upregulation of the genes responsible for FASN in prostate cancer mice. Moreover, inhibition of the enzyme limited the growth of the tumour cells. A characteristic up-regulation of frizzled class receptor 6 (FZD6) has been shown to suppress the development of prostate cancer cells, suggesting the involvement of the Wingless-related integration site (Wnt) signalling pathway in prostate cancer (Han et al., 2018). The findings suggested that suppressing Wnt signalling pathway might help to prevent disease progression.
TARGETING METABOLIC PATHWAY PROTEINS
Proteomic analysis using LC-MS/MS have revealed the involvement of the Wnt signalling pathway in the prostate. Wnt signalling through transient receptor potential melastatin member 8 (TRPM8) triggers the stemness of prostate cancer. The proteomic analysis of the cellular signalling mechanism revealed the participation of FZD6 in the signalling of the prostate tumour. Proliferation and migration of the tumours were shown to be inhibited by Luteolin, further confirming the participation of TRPM8 in the signalling (Asuthkar et al., 2016). A liquid chromatography analysis of the protein regulated by miR-200b confirmed the findings presented by Asuthkar et al. (2016). He et al. (2018) used prostate cancer cell lines to demonstrate the involvement of transmembrane 4 L6 family member 1 (TM4SF1), yes-associated protein 1 (YAP1), Protein phosphatase inhibitor 2 (PPP1R2), Myristoylated alanine-rich C-kinase substrate (MARCKS), and Reticulon 4 (RTN4) inhibitors in reducing the progression of prostate cancer. The inhibitors analysed targeted the Wnt pathway. Also, 13 proteins were found to be upregulated in miR-200b mimics (Park et al., 2017). These findings highlight the importance of proteomic analysis in understanding the cellular signalling of prostate cancer. The application of proteomics tools in the field of prostate cancer has opened exciting opportunities for the study of protein-inhibitor based therapeutic interventions. Currently, the majority of anti-prostate cancer drugs in clinical trials are based on cellular signalling inhibition (Xie et al., 2019).
The investigation of the metabolic pathways involved in the aetiology and progression of PCa has also provided insight into the mechanisms involved in the regulation of enzymes in the fatty acid biosynthetic pathways. For instance, LC/MS and Western Blot analysis in animal models have provided evidence of the dysregulation in the fatty acid biosynthetic pathway. FASN and monoacylglycerol lipase (MAGL) are enzymes which create pools of cellular fatty acids whereas fatty acid binding protein 5 (FABP5) is an intracellular chaperone which delivers fatty acids to nuclear receptors to augment PCa metastasis. As FABP5, FASN, and MAGL have been shown to be independently involved in PCa progression, the researchers hypothesised that FABP5 represents a central mechanism that links cytosolic lipid metabolism to pro-metastatic signalling of nuclear receptors. Examination of FABP5 function in mice has shown that the ability of MAGL and FASN to promote nuclear receptor activation and PCa metastasis depends on FABP5 expression (Figure 1) (Carbonetti et al., 2019).
Figure 1: FABP5 is required for in vivo prostate cancer metastasis mediated by FASN and MAGL. (A) Demonstrative luciferase signals of PC3-Luc cells expressing vector, MAGL, or FASN on days 7 (D7) and 49 (D49) respectively. Signals were seen for PC3-Luc cells that express FABP5 shRNA (shFABP5) as well as those expressing FABP5 shRNA while overexpressing FASN (FASN / shFABP5) or MAGL (MAGL / shFABP5). (B) Total vector and shFABP5 cell flux at the primary tumour site, whole mouse, and femurs. (C) Total vector flux, FASN, and FASN / shFABP5 cells at the primary tumour site, whole mouse, and femurs. (D) Total vector flux, MAGL, and MAGL / shFABP5 cells at the primary tumour site, whole mouse, and femurs. Data was given as means ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001; **** p < 0.0001 vs. vector. # p < 0.05; ## p < 0.01; ### p < 0.001; ####p < 0.0001 represents FASN vs. FASN/shFABP5 or MAGL vs. MAGL/shFABP5; (n = 8) (Carbonetti et al., 2019).
BIOMARKERS FOR PROSTATE CANCER
Pathway regulation through phosphorylation has also been used to enhance the understanding of the mechanisms of prostate cancer. Transcription factors are tightly regulated through phosphorylation pathways; thus, the application of phosphoproteomic datasets to analyse pathways may provide insights into the mechanism of prostate cancer. Drake et al. (2016) demonstrated the existence of patient unique signalling networks in patients with castration-resistant prostate cancer (CRPC). The researchers reported significant differences in signalling pathways among CRPC patients. The differences indicated by the researchers could be used to segregate prostate cancer patients into different clinical categories.
Furthermore, the identification of phosphorylation-dependent differences in the metastasis pathways could be used to provide personalized treatment to patients. The research into phosphorylation-dependent prostate cancer hallmarks offers a unique advantage to improve cancer medication by developing targeted therapies (Xu et al., 2016). However, it is essential to note that despite the promise to deliver more accurate and targeted treatment, the cost associated with these kind of tests are yet to be developed and require significant financial investment.
Cellular signalling networks have been used to develop biomarkers for cancer diagnosis. The advancement in molecular and proteomic techniques such as Quantitative Proteomics Signature Profiling (qPSP) and Paired Fuzzy SubNetworks (PFSNet), has enhanced network analysis in prostate cancer patients (Goh et al., 2019). In protein complexes, qPSP has been shown to increase the biological content of proteomic data by translating protein expressions into hit-rates. Additionally, qPSP can be used for the study of extremely heterogeneous clinical prostate cancer proteomics data, considering its capacity to differentiate phenotype groups even at small sample sizes and high noise robustness, as well as its improved summary statistics (Goh et al., 2015; Wang et al., 2017). Likewise, it has been observed that PFSNet-identified subnetworks have greater accuracy over independently collected datasets compared to other approaches (Lim and Wong, 2014). Via network-based methods such as PFSNet and qPSP, researchers have noticed that low overlaps in patient data can emerge from a lack of sensitivity of the analytical methodology used, when considering the apparent overwhelming amount of heterogeneity amongst patients with prostate cancer (Fischer et al., 2019; Ho et al., 2020). Goh et al. (2019), reported that network-based analysis is more effective than protein analysis approaches in prostate cancer diagnosis. In general, network-based analysis provides more accuracy, precision, and sensitivity in diagnosis (Goh et al., 2019).
THE POTENTIAL USE OF EXOSOMES AS BIOMARKERS
The analysis of biofluid has broadened the approaches applied to diagnose PCa. The involvement between extracellular vesicles in intracellular signalling provides a promising diagnostic strategy for PCa (Dhondt et al., 2020). According to Dhondt et al. (2020), these extracellular fluids can be used as biomarkers for diagnosis and prognosis of PCa. With the application of high throughput centrifugation and mass spectrometry techniques, extracellular vesicles have been extracted and analysed in urine from men with benign and malignant prostate tumours (Sequeiros et al., 2016; Dhondt et al., 2020). Further analysis of the proteome profile of the extracellular vesicles has helped to separate individuals with prostate cancer from those without the disease. The advances in proteome analysis of biofluids such as urine increase opportunities for diagnosis of the disease through non-invasive methods.
Extracellular vesicles are involved in intracellular communication where they carry a variety of cargo from the intracellular space to extracellular fluid. The vesicles may be involved in trafficking of cargo such as metabolites, non-coding RNA, and/or proteins (Nguyen et al., 2019). The molecular components of the extracellular vesicles can be used to examine the functionality of the parent cells, thus providing insight into the state of the cell (whether cancerous or not) (Nguyen et al., 2019; Hornung et al., 2020). The use of extracellular fluid to analyse the cellular state is a new technique that is currently being considered for diagnosis and prognosis of prostate cancer. Hornung et al. (2020), analysed the exosome composition of VCaP cells and reported a significant resemblance between the extracellular vesicles and the parent cells. The findings reaffirmed the applicability of exosome analysis as a useful proteomic tool for prostate cancer diagnosis (Ishizuya et al., 2020).
PROTEOME ANALYSIS ENHANCE PERSONALIZED TREATMENT
The use of biomarkers to enhance personalised prostate cancer treatment is rapidly gaining popularity. However, the majority of biomarkers currently available for cancer diagnosis and prognosis have not been validated and show poor reproducibility within a cohort of patients. This caveat hinders the application of these biomarkers in clinical settings. Thus, research into the identification of robust novel biomarkers focuses on finding more effective molecular signatures for disease diagnosis. Kim et al. (2016), used Multiplexed SRM-MS to assess the effectiveness of biomarkers for the diagnosis of prostate cancer in urine samples. Out of 48 urine samples analysed, the researchers identified ANXA3, IDHC, PEDF, PRDX6, SERA, and TGM4 as potential biomarkers for use in the diagnosis of prostate cancer (Jadvar et al., 2020).
Proteome analysis of the bones among prostate cancer patients has revealed a significant heterogeneity. An analysis of bone metastasis demonstrated that there are more diverse proteins participating in cell cycle regulation compared to primary prostate tumours. Molecular changes such as alteration of processing of RNA and damages in DNA were identified (Zhang et al., 2019). Metabolic changes were also observed, such as oxidation of fatty acids and carbohydrate metabolism. In general, these metabolic alterations in bone metastasis were associated with cellular adhesion characteristics of the bone cells. These findings were indicative of the possibility to apply bone metastasis as a diagnostic measure for prostate cancer (Rodríguez-Blanco et al., 2018).
Multiple studies have focused on the total cell protein alterations in prostate cancer in the identification of novel biomarkers (Welton et al., 2016; Totten et al., 2018; Sinha et al., 2019). Moreover, the researchers emphasised the effectiveness of affordable proteomic analysis tools such as mass spectrometry. Compared to the expensive and rare genetic tools, including DNA/RNA sequencing approaches, it is rational to apply proteomic analysis (Latonen et al., 2018). Furthermore, integrative proteomic analyses are more precise and accurate compared to molecular examination of DNA methylation and gene analysis. Latonen et al. (2018) suggested that the application of more effective proteome analysis methods could speed up the process of personalised prostate cancer treatment.
PROTEOMIC PROFILING OF PROSTATE CANCER
The total protein profile in prostate cancer tissues is an essential tool for diagnosis and monitoring of disease progression (Zhou et al., 2019). The understanding of global alteration in the cellular protein profile may hasten the discovery of robust diagnostic methods and personalized treatment. However, the knowledge of changes in the protein components of prostate cancer cells is still limited (Zhang et al., 2018). Quantitative proteomic analysis to profile the total protein constituents of prostate cancer cells has been performed using transgenic mice (Zhang et al., 2018). Zhang et al. (2018) compared the total protein constituted of transgenic mice against the protein profile of wild type mice and reported 61 proteins that were differentially expressed between wild type and transgenic mice. Researchers suggest that bioinformatics tools combined with accurate proteomic tools can help leverage the power of the proteome profile in cancerous cells to perform a precise diagnosis of prostate cancer (Pappireddi et al., 2019).
PROTEIN PROFILING AND PROSTATE CANCER DRUG DISCOVERY
Stelloo et al. (2018) analysed the androgen receptor protein profile using immunoassays. The researchers identified androgen receptor proteins among other interactors. Interestingly, most of the identified proteins were required for the progression of prostate cancer (Stelloo et al., 2018). Further analysis using chromatin immunoassays and sequence-based assays enabled the researchers to identify distinct sub-complexes of androgen receptor interactors. The complexes identified were selectively adapted to tumorigenesis. The findings reported by Stelloo et al. (2018) emphasised the importance of performing proteome analysis as a diagnostic approach for prostate cancer (Figure 2).
Figure 2: Clusters linked to carcinogenesis of prostate cancer were identified by application of both proteomic and genetic analysis. (A) Principal component analysis scores plot based on the read counts of each TF in AR-binding sites under R1881 conditions. (B) Heatmap of the Top 2000 regions with the most variable binding at AR sites of the TFs indicated. Based on the hierarchical clustering 3 clusters were identified. (C) Radar plot showing pattern enrichment in the Top 2000 variable regions (separated in the three clusters). The radii lengths are the absolute Z-score. The pattern colours correspond to the TF families. (D) Heatmap showing AR ChIP-seq signal (FPKM) in three clusters of LHSAR cells transduced with LacZ control, FOXA1, HOXB13 or both. Data is centred at AR peaks, depicting a 5-kb window around the peak. (E) Boxplot visualising the normalised AR signal (FPKM) at AR binding sites in clusters 1.1 (blue), 1.2 (red) and 1.3 (green). p < 0.05, * * * p < 1e−16 (t-test) (Stelloo et al., 2018).
Prostate cancer chemotherapy involving the inhibition of androgen receptors expression can be investigated in order to improve treatment outcomes. However, the inhibition of androgen receptor expression requires an alteration in heat shock protein expression—a process that involves the identification of useful biomarkers. Robust predictive biomarkers may contribute to the strategy of personalised treatment. Stelloo et al. (2018) applied MS analysis to the expression of heat shock protein 90 in order to discover how its inhibition could be used to inhibit the proliferation of prostate cancer cells. The researchers reported eight proteins that were inhibited that belonged to the cell cycle regulation family. The findings provided insight into the application of proteome profile analysis to identify putative protein targets for drug discovery. In addition, the protein inhibition profiles were characteristic to some patients, which suggested the possibility of personalised treatment. Other studies (Garrido-Rodríguez et al., 2019; Singh and Sharma, 2020) have also reported the role of SWATH protein profiling tool in unveiling new drug targets. It is, therefore, imperative to use the protein profiling tools for the identification of putative drug target proteins in the process of prostate cancer drug discovery.
Coupling microscopy and laser capture microdissection (LCM) techniques have enabled scientists to target and extract specific cell types from tissues. A combination of LCM and LC/MS-MS has been used to remove proteins from prostate tissue (Staunton et al., 2016). The researchers reported high accuracy in identification and isolation of the target cells. The findings from the literature have demonstrated the applicability of protein profiling in drug target identification to enhance the specificity of newly developed cancer drugs while limiting cytotoxicity effects to non-target cells.
CONCLUSION
This literature review has reported evidence of the physiological processes involved in the proliferation of prostate cancer cells. Metabolic signalling and regulation play a central role in the aetiology of prostate cancer. Aberrant regulation of lipid metabolism due to abnormal expression of FASN enzyme has been widely documented in the literature. An abnormal increase in the expression of this enzyme has been associated with prostate cancer. Prostate cancer diagnosis and prognosis has been enhanced through the use of biomarkers. The literature search showed that numerous biomarkers had been recommended in the diagnosis of prostate cancer. However, the challenge is that the majority of the biomarkers are specific, limiting their application to research only. The use of prostate cancer biomarkers has also been extended to biofluid analysis of extracellular vesicles. These vesicles have been shown to contain protein profiles similar to their parent cells, thus demonstrating the application of non-invasive diagnostic tools in the diagnosis of prostate cancer. The literature search on the application of proteomic profiling has revealed a high potential for enhancing drug targeting during the drug development process. Profiling of total proteins in prostate cancer cells will help the drug development process optimise drug activity while limiting cytotoxicity to non-target cells. Future studies should focus on the incorporation of robust bioinformatics tools for more effective drug modelling and target optimisation.
REFERENCES
Asuthkar, S., Demirkhanyan, L., Mueting, S. R., Cohen, A. and Zakharian, E. (2016) ‘High-throughput proteome analysis reveals targeted TRPM8 degradation in prostate cancer’, Oncotarget, 8(8), 12877-12890, available: https://doi.org/10.18632/oncotarget.14178.
Boja, E. S. and Rodriguez, H. (2014) ‘Proteogenomic convergence for understanding cancer pathways and networks’, Clinical Proteomics, 11(1), available: https://doi.org/10.1186/1559-0275-11-22.
Carbonetti, G., Wilpshaar, T., Kroonen, J., Studholme, K., Converso, C., d’Oelsnitz, S. and Kaczocha, M. (2019) ‘FABP5 coordinates lipid signaling that promotes prostate cancer metastasis’, Scientific Reports, 9(1), available: https://doi.org/10.1038/s41598-019-55418-x.
Dhondt, B., Geeurickx, E., Tulkens, J., Van Deun, J., Vergauwen, G., Lippens, L., Miinalainen, I., Rappu, P., Heino, J., Ost, P., Lumen, N., De Wever, O. and Hendrix, A. (2020) ‘Unravelling the proteomic landscape of extracellular vesicles in prostate cancer by density-based fractionation of urine’, Journal Of Extracellular Vesicles, 9(1), 1736935, available: https://doi.org/10.1080/20013078.2020.1736935.
Drake, J. M., Paull, E. O., Graham, N. A., Lee, J. K., Smith, B. A., Titz, B., Stoyanova, T., Faltermeier, C. M., Uzunangelov, V., Carlin, D. E., Fleming, D. T., Wong, C. K., Newton, Y., Sudha, S., Vashisht, A. A., Huang, J., Wohlschlegel, J.A., Graeber, T.G., Witte, O.N., and Stuart, J. M. (2016) ‘Phosphoproteome Integration Reveals Patient-Specific Networks in Prostate Cancer’, Cell, 166(4), pp.1041-1054, available: https://doi.org/10.1016/j.cell.2016.07.007.
Felici, A., Pino, M. S. and Carlini, P. (2012) ‘A Changing Landscape in Castration-Resistant Prostate Cancer Treatment’, Frontiers in Endocrinology, 3, available: https://doi.org/10.3389/fendo.2012.00085.
Fiñones, R. R., Yeargin, J., Lee, M., Kaur, A. P., Cheng, C., Sun, P., Wu, C., Nguyen, C., Wang-Rodriguez, J., Meyer, A. N., Baird, S. M., Donoghue, D. J. and Haas, M. (2013) ‘Early Human Prostate Adenocarcinomas Harbor Androgen-Independent Cancer Cells’, PLoS ONE, 8(9), p.e74438, available: https://doi.org/10.1371/journal.pone.0074438.
Fischer, S., Tahoun, M., Klaan, B., Thierfelder, K. M., Weber, M., Krause, B. J., Hakenberg, O., Fuellen, G. and Hamed, M. (2019) ‘A Radiogenomic Approach for Decoding Molecular Mechanisms Underlying Tumor Progression in Prostate Cancer’, Cancers, 11(9), p.1293, available: https://doi.org/10.3390/cancers11091293.
Garrido-Rodríguez, M., Ortea, I., Calzado, M. A., Muñoz, E. and García, V. (2019) ‘SWATH proteomic profiling of prostate cancer cells identifies NUSAP1 as a potential molecular target for Galiellalactone’, Journal of Proteomics, 193, pp.217-229, available: https://doi.org/10.1016/j.jprot.2018.10.012.
Goh, W. W. B., Guo, T., Aebersold, R. and Wong, L. (2015) ‘Quantitative proteomics signature profiling based on network contextualization’, Biology Direct, 10(1), available: https://doi.org/10.1186/s13062-015-0098-x.
Goh, W. W. B., Zhao, Y., Sue, A. C., Guo, T. and Wong, L. (2019) ‘Proteomic investigation of intra-tumor heterogeneity using network-based contextualization — A case study on prostate cancer’, Journal of Proteomics, 206, p.103446, available: https://doi.org/10.1016/j.jprot.2019.103446.
Han, K., Lang, T., Zhang, Z., Zhang, Y., Sun, Y., Shen, Z., Beuerman, R. W., Zhou, L. and Min, D. (2018) ‘Luteolin attenuates Wnt signaling via upregulation of FZD6 to suppress prostate cancer stemness revealed by comparative proteomics’, Scientific Reports, 8(1), available: https://doi.org/10.1038/s41598-018-26761-2.
He, M., Gou, M., Qi, M., Xiang, W., Ji, Z., Wang, W., Zhao, S. and Liu, Y. (2018) ‘Label free quantitative proteomics reveals the role of miR-200b in androgen-independent prostate cancer cells’, Clinical Proteomics, 15(1), available: https://doi.org/10.1186/s12014-018-9185-1.
Ho, S. Y., Wong, L. and Goh, W. W. B. (2020) ‘Avoid Oversimplifications in Machine Learning: Going beyond the Class-Prediction Accuracy’, Patterns, 1(2), p.100025, available: https://doi.org/10.1016/j.patter.2020.100025.
Hornung, T., O’Neill, H. A., Logie, S. C., Fowler, K. M., Duncan, J. E., Rosenow, M., Bondre, A.S., Tinder, T., Maher, V., Zarkovic, J., Zhong, Z., Richards, M. N., Wei, X., Miglarese, M. R., Mayer, G., Famulok, M. and Spetzler, D. (2020) ‘ADAPT identifies an ESCRT complex composition that discriminates VCaP from LNCaP prostate cancer cell exosomes’, Nucleic Acids Research, 48(8), pp.4013-4027, available: https://doi.org/10.1093/nar/gkaa034.
Iglesias-Gato, D., Wikström, P., Tyanova, S., Lavallee, C., Thysell, E., Carlsson, J., Hägglöf, C., Cox, J., Andrén, O., Stattin, P., Egevad, L., Widmark, A., Bjartell, A., Collins, C. C., Bergh, A., Geiger, T., Mann, M. and Flores-Morales, A. (2016) ‘The Proteome of Primary Prostate Cancer’, European Urology, 69(5), pp.942-952, available: https://doi.org/10.1016/j.eururo.2015.10.053.
Iglesias-Gato, D., Thysell, E., Tyanova, S., Crnalic, S., Santos, A., Lima, T. S., Geiger, T., Cox, J., Widmark, A., Bergh, A., Mann, M., Flores-Morales, A. and Wikström, P. (2018) ‘The Proteome of Prostate Cancer Bone Metastasis Reveals Heterogeneity with Prognostic Implications’, Clinical Cancer Research, 24(21), pp.5433-5444, available: https://doi.org/10.1158/1078-0432.CCR-18-1229.
Ishizuya, Y., Uemura, M., Narumi, R., Tomiyama, E., Koh, Y., Matsushita, M., Nakano, K., Hayashi, Y., Wang, C., Kato, T., Hatano, K., Kawashima, A., Ujike, T., Fujita, K., Imamura, R., Adachi, J., Tomonaga, T. and Nonomura, N. (2020) ‘The role of actinin-4 (ACTN4) in exosomes as a potential novel therapeutic target in castration-resistant prostate cancer’, Biochemical and Biophysical Research Communications, 523(3), pp.588-594, available: https://doi.org/10.1016/j.bbrc.2019.12.084.
Jadvar, H., Ballas, L. K., Choyke, P. L., Fanti, S., Gulley, J. L., Herrmann, K., Hope, T. A., Klitzke, A. K., Oldan, J. D., Pomper, M. G., Rowe, S. P., Subramaniam, R. M., Taneja, S. S., Vargas, H. A. and Ahuja, S. (2020) ‘Appropriate Use Criteria for Imaging Evaluation of Biochemical Recurrence of Prostate Cancer After Definitive Primary Treatment’, Journal of Nuclear Medicine, 61(4), pp.552-562, available: https://doi.org/10.2967/jnumed.119.240929.
Kim, Y., Jeon, J., Mejia, S., Yao, C. Q., Ignatchenko, V., Nyalwidhe, J. O., Gramolini, A. O., Lance, R. S., Troyer, D. A., Drake, R. R., Boutros, P.C., Semmes, O. J. and Kislinger, T. (2016) ‘Targeted proteomics identifies liquid-biopsy signatures for extracapsular prostate cancer’, Nature Communications, 7(1), available: https://doi.org/10.1038/ncomms11906.
Latonen, L., Afyounian, E., Jylhä, A., Nättinen, J., Aapola, U., Annala, M., Kivinummi, K. K., Tammela, T. T. L., Beuerman, R. W., Uusitalo, H., Nykter, M. and Visakorpi, T. (2018) ‘Integrative proteomics in prostate cancer uncovers robustness against genomic and transcriptomic aberrations during disease progression’, Nature Communications, 9(1), available: https://doi.org/10.1038/s41467-018-03573-6.
Li, F., Glinskii, O. V., Mooney, B. P., Rittenhouse-Olson, K., Pienta, K. J. and Glinsky, V. V. (2017) ‘Cell surface Thomsen-Friedenreich proteome profiling of metastatic prostate cancer cells reveals potential link with cancer stem cell-like phenotype’, Oncotarget, 8(58), available: https://doi.org./10.18632/oncotarget.21985.
Lim, K. and Wong, L. (2014) ‘Finding consistent disease subnetworks using PFSNet’, Bioinformatics, 30(2), pp.189-196, available: https://doi.org/10.1093/bioinformatics/btt625.
Mann, M., Kulak, N. A., Nagarjuna, N. and Cox, J. (2013) ‘The Coming Age of Complete, Accurate, and Ubiquitous Proteomes’, Molecular Cell, 49(4), pp.583-590, available: https://doi.org/10.1016/j.molcel.2013.01.029.
Murray, H. C., Dun, M. D. and Verrills, N. M. (2017) ‘Harnessing the power of proteomics for identification of oncogenic, druggable signalling pathways in cancer’, Expert Opinion on Drug Discovery, 12(5), pp.431-447, available: https://doi.org/10.1080/17460441.2017.1304377.
Nguyen, E. V., Centenera, M. M., Moldovan, M., Das, R., Irani, S., Vincent, A. D., Chan, H., Horvath, L. G., Lynn, D. J., Daly, R. J. and Butler, L. M. (2018) ‘Identification of Novel Response and Predictive Biomarkers to Hsp90 Inhibitors Through Proteomic Profiling of Patient-derived Prostate Tumor Explants’, Molecular & Cellular Proteomics, 17(8), pp.1470-1486, available: https://doi.org/10.1074/mcp.RA118.000633.
Nguyen, E. V., Pereira, B. A., Lawrence, M. G., Ma, X., Rebello, R. J., Chan, H., Niranjan, B., Wu, Y., Ellem, S., Guan, X., Wu, J., Skhinas, J. N., Cox, T. R., Risbridger, G. P., Taylor, R. A., Lister, N. L. and Daly, R. J. (2019) ‘Proteomic Profiling of Human Prostate Cancer-associated Fibroblasts (CAF) Reveals LOXL2-dependent Regulation of the Tumor Microenvironment’, Molecular & Cellular Proteomics, 18(7), pp.1410-1427, available: https://doi.org/10.1074/mcp.RA119.001496.
Office for National Statistics. (2019) Cancer Registration Statistics, England: 2017. London: Government Statistical Service (GSS), pp.1-4, available: https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/bulletins/cancerregistrationstatisticsengland/2017.
Pappireddi, N., Martin, L. and Wühr, M. (2019), ‘A Review on Quantitative Multiplexed Proteomics.’, ChemBioChem, 20(10), pp.1210-1224, available: https://doi.org/10.1002/cbic.201800650.
Park, A., Lee, J., Mun, S., Kim, D. J., Cha, B. H., Moon, K. T., Yoo, T. K. and Kang, H. (2017) ‘Identification of Transcription Factor YY1 as a Regulator of a Prostate Cancer-Specific Pathway Using Proteomic Analysis’, Journal of Cancer, 8(12), pp.2303-2311, available: https://doi.org/10.7150/jca.19036.
Rodríguez-Blanco, G., Zeneyedpour, L., Duijvesz, D., Hoogland, A. M., Verhoef, E. I., Kweldam, C. F., Burgers, P. C., Smitt, P. S., Bangma, C. H., Jenster, G., van Leenders, G. J. L. H., Dekker, L. J. M. and Luider, T. M. (2018), ‘Tissue proteomics outlines AGR2 AND LOX5 as markers for biochemical recurrence of prostate cancer’, Oncotarget, 9(92), available: https://doi.org/10.18632/oncotarget.26342.
Schubert, O. T., Röst, H. L., Collins, B. C., Rosenberger, G. and Aebersold, R. (2017) ‘Quantitative proteomics: challenges and opportunities in basic and applied research’, Nature Protocols, 12(7), pp.1289-1294, available: https://doi.org/10.1038/nprot.2017.040.
Sequeiros, T., Rigau, M., Chiva, C., Montes, M., Garcia-Grau, I., Garcia, M., Diaz, S., Celma, A., Bijnsdorp, I., Campos, A., Di Mauro, P., Borrós, S., Reventós, J., Doll, A., Paciucci, R., Pegtel, M., de Torres, I., Sabidó, E., Morote, J. and Olivan, M. (2016) ‘Targeted proteomics in urinary extracellular vesicles identifies biomarkers for diagnosis and prognosis of prostate cancer’, Oncotarget, 8(3), available: https://doi.org/10.18632/oncotarget.13634.
Singh, A. N. and Sharma, N. (2020). ‘Quantitative SWATH-Based Proteomic Profiling for Identification of Mechanism-Driven Diagnostic Biomarkers Conferring in the Progression of Metastatic Prostate Cancer’, Frontiers in Oncology, 10, available: https://doi.org/10.3389/fonc.2020.00493.
Sinha, A., Huang, V., Livingstone, J., Wang, J., Fox, N. S., Kurganovs, N., Ignatchenko, V., Fritsch, K., Donmez, N., Heisler, L. E., Shiah, Y. J., Yao, C. Q., Alfaro, J. A., Volik, S., Lapuk, A., Fraser, M., Kron, K., Murison, A., Lupien, M., Sahinalp, C., Collins, C. C., Tetu, B., Masoomian, M., Berman, D. M., van der Kwast, T., Bristow, R. G., Kislinger, T. and Boutros, P. C. (2019) ‘The Proteogenomic Landscape of Curable Prostate Cancer’, Cancer Cell, 35(3), pp.414-427.e6, available: https://doi.org/10.1016/j.ccell.2019.02.005.
Staunton, L., Tonry, C., Lis, R., Finn, S., O-Leary, J., Loda, M., Bowden, M. and Pennington, S. R. (2016) ‘Profiling the tumor microenvironment proteome in prostate cancer using laser capture microdissection coupled to LC-MS-A technical report’, EuPA Open Proteomics, 10, pp.19-23, available: https://doi.org/10.1016/j.euprot.2015.11.001.
Stelloo, S., Nevedomskaya, E., Kim, Y., Hoekman, L., Bleijerveld, O. B., Mirza, T., Wessels, L. F. A., van Weerden, W. M., Altelaar, A. F. M., Bergman, A. M. and Zwart, W. (2018) ‘Endogenous androgen receptor proteomic profiling reveals genomic subcomplex involved in prostate tumorigenesis’, Oncogene, 37(3), pp.313-322, available: https://doi.org/10.1038/onc.2017.330.
Testa, U., Castelli, G. and Pelosi, E. (2019) ‘Cellular and Molecular Mechanisms Underlying Prostate Cancer Development: Therapeutic Implications’, Medicines, 6(3), p.82, available: https://doi.org/10.3390/medicines6030082.
Totten, S. M., Adusumilli, R., Kullolli, M., Tanimoto, C., Brooks, J.D., Mallick, P. and Pitteri, S. J. (2018) ‘Multi-lectin Affinity Chromatography and Quantitative Proteomic Analysis Reveal Differential Glycoform Levels between Prostate Cancer and Benign Prostatic Hyperplasia Sera’, Scientific Reports, 8(1), available: https://doi.org/10.1038/s41598-018-24270-w.
Wang, G., Zhao, D., Spring, D. J. and DePinho, R. A. (2018) ‘Genetics and biology of prostate cancer’, Genes & Development, 32(17-18), pp.1105-1140, available: https://doi.org/10.1101/gad.315739.118.
Wang, W., Sue, A. C. and Goh, W.W.B. (2017) ‘Feature selection in Clinical Proteomics: with great power comes great reproducibility’, Drug Discovery Today, 22(6), pp.912-918, available: https://doi.org/10.1016/j.drudis.2016.12.006.
Welton, J. L., Brennan, P., Gurney, M., Webber, J. P., Spary, L. K., Carton, D. G., Falcón-Pérez, J. M., Walton, S. P., Mason, M. D., Tabi, Z. and Clayton, A. (2016) ‘Proteomics analysis of vesicles isolated from plasma and urine of prostate cancer patients using a multiplex, aptamer-based protein array’, Journal Of Extracellular Vesicles, 5(1), p.31209, available: https://doi.org/10.3402/jev.v5.31209.
Wyatt, A. W., Mo, F., Wang, K., McConeghy, B., Brahmbhatt, S., Jong, L., Mitchell, D. M., Johnston, R. L., Haegert, A., Li, E., Liew, J., Yeung, J., Shrestha, R., Lapuk, A. V., McPherson, A., Shukin, R., Bell, R. H., Anderson, S., Bishop, J., Hurtado-Coll, A., Xiao, H., Chinnaiyan, A. M., Mehra, R., Lin, D., Wang, Y., Fazli, L., Gleave, M. E., Volik, S. V. and Collins, C .C. (2014) ‘Heterogeneity in the inter-tumor transcriptome of high risk prostate cancer’, Genome Biology, 15(8), available: https://doi.org/10.1186/s13059-014-0426-y.
Xie, J., Zhang, A. H., Qiu, S., Zhang, T. L., Li, X. N., Yan, G. L., Sun, H., Liu, L. and Wang, X. J. (2019) ‘Identification of the perturbed metabolic pathways associating with prostate cancer cells and anticancer affects of obacunone’, Journal of Proteomics, 206, p.103447, available: https://doi.org/10.1016/j.jprot.2019.103447.
Xu, G., Zou, W., Du, S., Wu, M., Xiang, T. and Luo, Z. (2016) ‘Mechanism of dihydroartemisinin-induced apoptosis in prostate cancer PC3 cells: An iTRAQ-based proteomic analysis’, Life Sciences, 157, pp.1-11, available: https://doi.org/10.1016/j.lfs.2016.05.033.
Ylipää, A., Kivinummi, K., Kohvakka, A., Annala, M., Latonen, L., Scaravilli, M., Kartasalo, K., Leppänen, S., Karakurt, S., Seppälä, J., Yli-Harja, O., Tammela, T. L. J., Zhang, W., Visakorpi, T., Nykter, M. (2015) ‘Transcriptome Sequencing Reveals PCAT5 as a Novel ERG-Regulated Long Noncoding RNA in Prostate Cancer’, Cancer Research, 75(19), 4026-4031, available: https://doi.org/10.1158/0008-5472.can-15-0217.
Yuan, W., Zhang, Z., Dai, B., Wei, Q., Liu, J., Liu, Y., Liu, Y., He, L. and Zhou, D. (2016) ‘Whole-exome sequencing of duodenal adenocarcinoma identifies recurrent Wnt/β-catenin signaling pathway mutations’, Cancer, 122(11), pp.1689-1696, available: https://doi.org/10.1002/cncr.29974.
Zhang, W., Hu, C., Wang, X., Bai, S., Cao, S., Kobelski, M., Lambert, J. R., Gu, J. and Zhan, Y. (2019) ‘Role of GDF15 in methylseleninic acid-mediated inhibition of cell proliferation and induction of apoptosis in prostate cancer cells’, PLoS ONE, 14(9), p.e0222812, available: https://doi.org/10.1371/journal.pone.0222812.
Zhang, Y., Wang, D., Li, M., Wei, X., Liu, S., Zhao, M., Liu, C., Wang, X., Jiang, X., Li, X., Zhang, S., Bergquist, J., Wang, B., Yang, C., Mi, J. and Tian, G. (2018) ‘Quantitative Proteomics of TRAMP Mice Combined with Bioinformatics Analysis Reveals That PDGF-B Regulatory Network Plays a Key Role in Prostate Cancer Progression’, Journal of Proteome Research, 17(7), pp.2401-2411, available: https://doi.org/10.1021/acs.jproteome.8b00158.
Zhou, B., Yan, Y., Wang, Y., You, S., Freeman, M. R. and Yang, W. (2019) ‘Quantitative proteomic analysis of prostate tissue specimens identifies deregulated protein complexes in primary prostate cancer’, Clinical Proteomics, 16(1), available: https://doi.org/10.1186/s12014-019-9236-2.