Author: Rebecca Krimmel
Institution: Louisiana State University
Date: July 2007
Abstract
The complete formation of nitrogenase is an essential part in understanding how the complex enzyme works and is essential for further manipulation and research of the structure. The exact formation process has yet to be determined, but it is hypothesized that models, containing gene deletions, derived using X-ray crystallographic analyses of nitrogenase structures, can provide snapshots of the formation process depending upon the deletion. To begin analysis of the formation, two types of nitrogenase were compared to each other from the Scissor Theory formation of nitrogenase (hypothesized in our laboratory), the nifB, cofactorless protein (cp) and the normal, holoprotein (hp) to determine whether or not the real structures matched with the Scissor Theory. The residues and p-cluster of the cp and hp, which make up the scissor structure, were located, isolated, and measured using the PyMOL computer program. Each residue and the p-cluster were looked at individually and as part of the complete structure. After using the pictures and measurements collected, it was determined the structures do follow the Scissor Theory. Now the distance, that the residues (around the cofactor) move will be taken, and the ratio will be applied to the handles. The ratio of the handles will allow for the continuation of this investigation to determine the structure of the nifH, FeS clusterless protein and to ultimately prove the Scissor Theory formation of nitrogenase. The results thus far indicate that the Scissor Theory may in fact explain the steps in the formation of nitrogenase.
Introduction
Nitrogenase is the most important enzyme used in nitrogen fixation, a process by which atmospheric nitrogen is converted to ammonia and can be summed up by the equation, N2 + 8 e- + 16 ATP + 8 H+ à 2 NH3 + 16 ADP + 16 Pi + H2 (Igarashi and Seefeldt 2003). This process allows plants to absorb the general atmospheric nitrogen into their roots, which ultimately lets other enzymes use it for the production of amino acids and other organic compounds (Burgess and Lowe 1996). These processes indirectly influence crop production in terms of the amount of protein that the plants contain. If the secrets behind nitrogenase are unlocked, the quality of crops potentially has the power to be greatly increased. A deeper understanding (than currently) of nitrogenase would provide more information for creating super-enriched protein based plants or other altered crops, providing hope for ending world hunger.
The structures of the holoprotein, normal structure of nitrogenase along with the nifB, cofactorless structure have been extensively studied. Research has verified changes in the arrangement of the residues around the cofactor in the nifB structure, which are expanded, compared to that of the holoprotein structure to prove that the final step in nitrogen formation is the insertion of the cofactor (Schmid, B et al. 2002; Zexing Cao, XJ and Q Zhang 2005). Other studies such as a density functional study of the cofactor have been used to gain insight to how the structures contain differences besides their obvious structural differences (Zexing Cao, XJ and Q Zhang 2005). Though, research has yet to determine the exact formation process of nitrogenase; the primary goal of nitrogenase research has been to determine the function and gain an extensive understanding of the processes involved with nitrogenase so the research has not focused on how the enzyme comes together. If the formation of nitrogenase is unlocked, it would open the pathway for scientists to explore how to better manipulate the creation of nitrogenase under laboratory settings.
article_1118_order_0
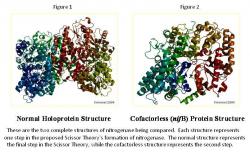
The exact process of the formation of nitrogenase is currently under investigation, with the pieces slowly coming together by utilizing known deletions in its genetic code. We hypothesize that by observing the structure of nitrogenase with different deletions, it is possible to predict the steps in the formation of the enzyme, culminating in the Scissor Theory, which is what is being analyzed. Certain forms of the molybdenum nitrogenase structure have been recorded and listed in the Protein Data Bank (PDB), while others are not and are still under investigation. The normal, holoprotein structure (Figure 1) is known, along with the nifB, cofactorless version of the structure of molybdenum nitrogenase (Figure 2); however, the nifH structure of nitrogenase is not known, which is missing the FeS cluster (Rubio and Ludden 2002). Each of these structures represents a step in the Scissor Theory formation of nitrogenase. If it is found that the known structures of nitrogenase fit into the Scissor Theory, it may be possible to create the third image, nifH structure, from a ratio of the opening found in the nifB structure. This research is analyzing the nifB and holoprotein structures to determine whether or not the structures fit the Scissor Theory and if so, the results will provide a starting point for the creation of the nifH structure. This research is extremely important because if the nitrogenase formation is solved, it will enable the scientific community to greatly further their research of the enzyme.
The Scissor Theory
article_1118_order_1
Three variations of the enzyme are key elements in determining exactly how and in what order the cofactor and FeS cluster are created in the molybdenum nitrogenase. The Scissor Theory, which contains the steps of formation of nitrogenase (hypothesized in our laboratory) (Figure 3), is where the cofactor and FeS cluster are inserted based on a pair of scissors. The tip of the scissors being the cofactor, the blades being residues, the fulcrum at which the blades and handles move being the p-cluster, the handles being residues connected to the p-cluster, and the FeS cluster being the unit between the handles that keep them closed or rather the safety lock on the scissors' handle. The proposed formation of nitrogenase starts with the nifH protein with the FeS cluster being inserted between the figurative handles, which pulls the handles together opening up the blades of the scissors for the cofactor to be inserted, as in the nifB protein, which closes as the complete structure is formed, as in the holoprotein. To determine whether or not nitrogenase is formed in this way, two of these three structures (those structures known) have been analyzed by comparing the residues and p-cluster in each structure of the normal and cofactorless variations.
article_1118_order_2
In the normal structure (Figure 4), the scissors appear completely closed. Everything is in place, the FeS cluster, p-cluster, and cofactor, while in the cofactorless structure (Figure 5), everything is closed and in place except for the cofactor, which has a space, such as the blades of scissors being open. Knowing that the fulcrum present in scissors allows for the space between the blades of the scissors to be proportional to the space between the handles, it is possible to predict the distance that will be open, when everything is closed and in place except for the FeS cluster that pulls both parts of the handle together. It is known that the nifH structure lacks the FeS cluster, which enables the future prediction of its structure using the information from the other structures, along with the Scissors Theory.
article_1118_order_3
This investigation is of the comparison of scissor structures of the normal and cofactorless proteins, which will ultimately allow the discovery of the nifH protein structure, thus completing the Scissor Theory and the understanding of the formation of nitrogenase.
Methods
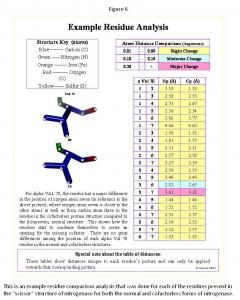
Using the Scissors Theory as a model, the amino residues of the scissors were identified and isolated in the nifB and holoprotein. First the cofactor, p-cluster, and all the residues within five angstroms of each were found in the PyMOL computer software. Next the residues connecting the cofactor and p-cluster were traced. Finally the residues, which make up the handles, surrounding the p-cluster were traced. Once the full scissor structure was isolated, all the components of the scissor structure were compared in the nifB and holoprotein structures with the structural changes noted. The method for the residue analysis (Figure 6) was created specifically for this project. Every residue's image was saved, and every atom in each residue was numbered and measured to all the other atoms present in its residue. This system showed all the changes present even very minor changes, subsequently a system of the degree of change was implemented (Figure 6), indicating "slight change" for 0.01 to 0.09 Angstroms difference, "moderate change" for 0.10 to 0.19 Angstroms difference, or "major change" for 0.20 and more Angstroms difference between the same residue points in each structure. This system highlighted the major differences to quickly determine the complete structural changes of each residue comparison. Once all the residues were analyzed, each component of the scissors ("handles," cofactor, p-cluster, and "blades") was separated, and the total structural changes were observed. In most cases the observations made about structure are reflected in the measurements. This provided means for determining whether the nifB and holoprotein structures followed the Scissor Theory.
Results
Note that the analyses of the Scissor Theory have been addressed as four general components, p-cluster, "blades," and cofactor, and the "handles," while the individual ligands' analyses have been omitted due to space.
article_1118_order_4
article_1118_order_5
The P-Cluster Comparison
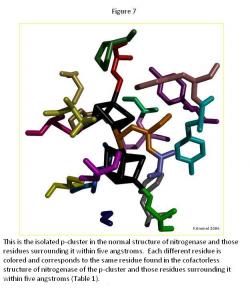
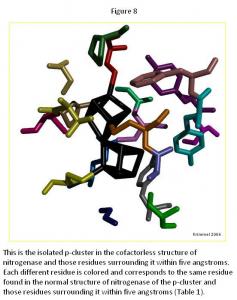
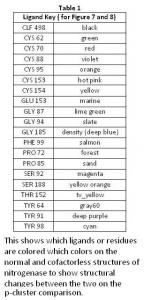
In the p-cluster, CLF 498, and surrounding ligands, many differences are seen between the normal structure (Figure 7) and the cofactorless structure (Figure 8) (Table 1). The p-cluster, which resides in the center, has five ligands (CYS 62, CYS 70, CYS 88, CYS 95, & CYS 153) bonded to it in the normal structure, but in the cofactor deficient structure there are only three (CYS 70, CYS 95, & CYS 154). Two of those three ligands (CYS 70 & CYS 95) are also bonded to the p-cluster in the normal structure. The third ligand is one that was not bonded to the p-cluster in the normal structure.
The remaining ligands, GLU 153, GLY 87, GLY 94, GLY 185, PHE 99, PRO 72, PRO 85, THR 152, TYR 64, and TYR 98, remain the same in form and position around the p-cluster except for SER 92, SER 188, and TYR 91. SER 92 moves to the opposite position it occupies in the normal structure and connects with TYR 91 on the other side of the p-cluster in the cofactor deficient structure. SER 188 keeps its general shape and form in both structures, but its oxygen atom moves towards the p-cluster in the normal structure.
Overall there is very little change in the two p-cluster structures, when compared to the cofactor in its normal and cofactor deficient conditions.
article_1118_order_6
article_1118_order_12
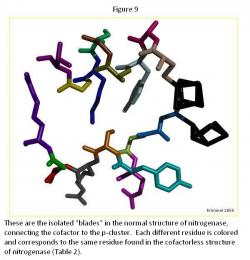
The "Blades" Comparison
The generally difference between the normal strand structure (Figure 9) and the cofactorless strand structure (Figure 10) (Table 2) is the fact that as the strand gets closer to the cofactor (or space the cofactor would occupy) in the cofactorless structure, each ligand compresses its structure, as compared to the normal structure, which allows for the hypothesis that the ligands await the arrival of the cofactor before closing up the opening.
article_1118_order_7
article_1118_order_13
The major difference is the ligand Arg 96, which is curled in the cofactor deficient structure creating more room for the cofactor, if it was in fact there. It generally curls toward the p-cluster in an attempt to create a window for the cofactor.
Throughout the ligands in the each strand, there was a general trend of the oxygen atoms moving inward towards the ligand, condensing each one, which allow for creating the cofactor window.
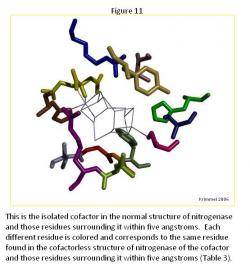
The Cofactor Comparison
There are major differences between the normal structure (Figure 11) and cofactorless structures (Figure 12) (Table 3). The first major difference between the two is the larger amount of space that the ligands of the cofactor deficient structure enclose as compared to the same space that the cofactor in the normal structure occupies. The cofactorless structure has a chain of ligands, which when the cofactor is not present, pulls away from the center.
article_1118_order_8
article_1118_order_9
The chain located below the cofactor contains four ligands, ARG 359, LEU 358, GLY 357, GLY 356, and ILE 355. In the normal structure, these ligands seem to as though they are the "stand" for the cofactor, while in the cofactor deficient structure; they unravel themselves, mainly by curling the ends and turning away from the cofactor space completely.
Another major difference is two ligands that are present within in the normal structure, HCA 494 and PHE 381, are not present in the cofactorless structure within 5 Angstroms of the cofactor space. These two ligands surround the cofactor and add to the general structure. HCA 494 connects to the cofactor adding more stability to the cofactor in the normal structure.
article_1118_order_10
article_1118_order_11
The other ligands, which are present towards the top of the cofactor space, CYS 275 (connected to the cofactor when present), SER 278, HIS 195, and VAL 70, remain relatively the same form in both structures as well as remaining in the same space in each structure.
The remaining ligands each changed to a noticeable degree from the other structure. In TYR 229, the tail turns towards the cofactor when it is present. In HIS 442, the ligand is connected to the cofactor when present, but when it is not present, the ring of the ligand moves towards the top of the structure and towards ARG 96. In ARG 96, the ligand curls and condenses when the cofactor is not present, but in general keeps the same position in both structures.
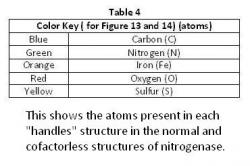
The "Handles" Comparison
There were no great differences among the ligands that intertwined to make up the "handles" in the normal structure (Figure 13) and in the cofactorless structure (Figure 14) (Table 4). They do alter slightly, not enough to change the Scissor Theory, but rather both structures are alike enough to support the Scissor Theory. The general structure of each remaining the same verify the "handles".
article_1118_order_14
article_1118_order_15
Discussion
The nifB and holoprotein forms of nitrogenase are important in discovering if the Scissor Theory is the method of the formation of nitrogenase. If the nifB structure proves to be "scissors" with the "blades" open and the holoprotein proves to be a pair of closed scissors, it can be determined that the structures follow the Scissor Theory. From the results obtained, each structure does in fact follow the Scissor Theory. There was a definite opening for the cofactor to be inserted in the nifB, which was not present in the holoprotein. Both structures follow the hypothetical pair of "scissors," the holoprotein with its structure closed and the nifB with its structure opened at the tip of the "blades." This is important because it proves two of the three steps in the Scissor Theory's formation of nitrogenase.
However, the residues surrounding the cofactor in the nifB structure provide inadequate information for determining a simple ratio to use for constructing the nifH structure, the last step to be determined in proving the Scissor Theory. With the open "blades" in the nifB structure, there is no distinct ratio between single residues. A combination of all the residues in the opened "blades" is what would be needed to determine the ratio for the nifH structure. Since PyMOL can only take measurements of atom to atom, there is no way of determining this ratio unless a pool of measurements is taken and an average ratio is determined so it could be applied to the "handles." Even though the nifH structure is future research, it is essential in continuing with the research of the Scissor Theory because the nifH structure is the final component needed to prove the entire Scissor Theory, and currently, the nifH structure does not exist in a X-ray crystallographic form to be observed. The results show no clear cut ratio that could be used to form the structure of the open "handles," but the structure of the nifB and holoprotein do follow the Scissor Theory.
Using the stated procedures for this study, there was no specific sources of error. The crystallographic structures were examined, and the exact structures were manipulated in PyMOL to get the measurements. Nonetheless the difference observed in the p-cluster in both structures is due to the different oxidation states of the structure and have no effect on the proposed Scissor Theory (Schmid, B et al. 2002).
Conclusion
After complete comparison analysis of the "blades," "fulcrum," and "handles, it was determined that the holoprotein and cofactorless protein do follow the Scissor Theory. The "fulcrum," and "handles" remained in the same in both structures, reinforcing the Scissor Theory. The nifB, cofactorless structure when compared to the normal structure, showed an opening in the "blades" and ligands around the cofactor, which also follow the Scissor Theory.
Future Investigations
Since the opening has been found of the "blades," the next step is to find the distance that the "blades" of the scissors open in the cofactorless protein and using that distance as a ratio for the "handles," to construct the nifH, FeS clusterless, protein structure.
Acknowledgements
RK would like to acknowledge God for this research opportunity, Dr. Brian Hales, Marcia Cotton, and Kandace Thomas for helping with everything that was needed, and the Office of Strategic Initiatives at LSU, LA-STEM, and the National Science Foundation for their support.
References
Burgess, BK and DJ Lowe (1996) Mechanism of Molybdenum Nitrogenase. Chemical
Reviews 96: 2983 3011.
Igarashi, RY and LC Seefeldt (2003) Nitrogen Fixation: The Mechanism of the Mo-Dependent
Nitrogenase. Critical Reviews in Biochemistry and Molecular Biology 38: 351 384.
Rubio, LM and PW Ludden. (2002) Nitrogen Fixation at the Millennium. Elsevier Science B. V.
Schmid, B et al. (2002) Structure of a Cofactor-Deficient Nitrogenase MoFe Protein. Science 296: 352 356.
Zexing Cao, XJ and Q Zhang. (2005) Density Functional Study of the Structure of the FeMo cofactor with an
Interstitial Atom and Homocitrate Ligand Ring Opening. Journal of Theoretical and Computational Chemistry 4: 593 602.