Authors: Prasanth Subramani, Premnath Rajakannu, Padhmanand Sudhakar, Naveen Jayaprakash
Institution: St. Peter's Engineering College, Anna University, Tamil Nadu, India
Date: December 2005
Abstract
Acquired immuno deficiency syndrome is a disease that manifests itself as a result of gene silencing and changing gene patterns caused by human immuno virus infection. Inspite of sustained efforts by the scientific community, a permanent treatment method for this infection has not been formulated till date. With the advancements in stem cell research, animal cell culture and genetic engineering, it is possible to devise a treatment for human immuno virus Infection. This article gives an insight into the convergence of the above mentioned technologies to yield a technique called Chromosome Engineering', which might be a viable treatment option for human immuno virus and other retroviral infections. We propose the insertion of the genes encoding for viral glycoprotein120, antisense strands of reverse transcriptase, integrase and protein27 into the eukaryotic vector Human Artificial Chromosome' to derive the novel therapeutic chromosome. The Engineered Therapeutic Chromosome can be introduced into Haematopoietic stem cells by micromanipulation or lipofection. These engineered cells can then be cultured in-vitro and introduced into the patients bone marrow. The expression of the inserted genes in the engineered cells has antiretroviral characteristics, thereby interrupting the HIV life cycle. Future research should focus on this technique so that it can be extended to clinical trials and may prove to be a potent treatment option against HIV and other retroviral infections.
Introduction
Figure 1. Life cycle of HIV-1.
Acquired immuno deficiency syndrome is the chronic stage of HIV-1 infection. The HIV-1 infection process is a complex mechanism involving the interaction of many viral and host proteins. The glycoprotein gp120 of Human Immuno Virus (HIV) attaches to the CD4 (Cluster of Differentiation) receptor of the T-Lymphocyte. The interaction between the envelope protein gp41 with transmembrane core receptors CXCR4' and CCR5' leads to fusion of viral and host cell membranes, leading to the entry of viral genome into the host cell. Virus-encoded reverse transcriptase coverts ssRNA into dsDNA while integrase causes the integration of viral genome with the host genome. Integration causes disruption in cytokine production and also uses the host cells for the production of viral particles causing further infection by cell lysis. Figure 1 shows the diagrammatic representation (Fauci 1988).
Conventional Treatment Strategies Classes of Anti-Retroviral Drugs
Table 1. Current drugs used to treat HIV-1 and their adverse effects.
Anti-retroviral drugs act by inhibiting the multiplication of the virus. Currently available drugs target two key enzymes the virus requires to multiply: Protease and Reverse Transcriptase. Drugs inhibiting the Reverse Transcriptase enzyme are divided into two types: Nucleoside Reverse Transcriptase Inhibitors (NRTIs) and Non-Nucleoside Reverse Transcriptase Inhibitors (NNRTIs). Table 1 lists the drugs currently used and their side effects.
Clinical reviews have shown that pharmacological treatment for HIV are toxic to vascular, neuronal, nephral and other systems, and are less potent than other therapies (e.g., McNicholl 2005, Moyle 2001, Mulligan et al. 2000).
Gene Therapy
Gene therapy is a technique for correcting defective genes responsible for disease development. It allows the insertion of anti-retroviral gene sequences into the patient's genome providing a treatment for HIV infection (Stephenson 1998).
Approaches include: inserting a normal gene into a nonspecific location in the genome to replace a nonfunctional gene; swapping a normal for abnormal gene through homologous recombination; repairing an abnormal gene through selective reverse mutation; and altering the regulation of a particular gene.
The technique, however, has many disadvantages:
Short-Lived Nature
The therapeutic DNA introduced into target cells must remain functional and the cells containing these must be long-lived and stable. Problems with integrating these into the genome and the rapidly dividing nature of many cells prevent gene therapy from achieving long-term benefits.
Immune Response
Therapeutic genes introduced into human tissues will evoke an immune response. If gene therapy has to be repeated, antigenic memory will induce a heightened immune response, which makes the therapy ineffective.
Problems with Viral Vectors
Viruses, though the carrier of choice, present a number of potential problems to the patient-toxicity, immune and inflammatory responses, and gene control and targeting issues and the fear that the vector, may recover its ability to cause disease.
Multigene Disorders
Conditions or disorders arising from single gene mutations are the best candidates for gene therapy. However, some most commonly occurring disorders, such as heart disease, high blood pressure, Alzheimer's disease, arthritis, and diabetes, are caused by the combined variation effects in many genes which would be more difficult to treat effectively using gene therapy.
With these disadvantages, the application of this treatment method is limited (Brenner 1995, Ali et al. 1994). Hence, we suggest that the chromosome engineering' technique may be best to provide a permanent treatment option against HIV infection.
The Chromosome Engineering Technique
Engineered chromosomes are inserted into Haematopoietic stem cells, which are stem cells capable of differentiating into various blood and immune cells. Since these engineered stem cells can differentiate into CD4+ T lymphocytes (which are the primary targets for HIV-1 infection), these cell lines are the most ideally suited for this therapeutic approach. These engineered stem cells can then be delivered into the patient by transfusion.
Figure 2. HAC (greenish red in color).
Human Artificial Chromosome: The Carrier
Human Artificial Chromosome (HAC) is an artificially constructed eukaryotic vector (Harrington et al. 1997, Ikeno et al. 1998), derived from Yeast Artificial Chromosome (YAC) (Henning et al. 1999). The essential elements of HAC are the centromere, which is a key element that ensures equal segregation of genetic material during cell division (Schueler et al. 2001), the replication centre that ensures faithful and stable DNA duplication, the telomeres which prevent the recombination of non homologous chromosomes. The telomeres of humans consist of as many as 2000 repeats of the sequence (Riethman et al. 2001). Figure 2 shows the Human Artificial Chromsome (HAC).
5'...TTAGGG TTAGGG TTAGGG TTAGGG TTAGGG TTAGGG..3'
3'...AATCCC AATCCC AATCCC AATCCC AATCCC AATCCC..5'
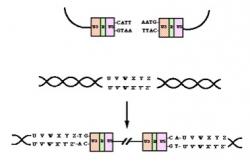
Construction of Hypothetical Therapeutic Chromosome
Figure 3. Model of Engineered HAC.
Computational analysis of the HIV-1 Genome shows the nucleotide sequences of gp120, reverse transcriptase and integrase enzyme systems (NCBI 2005). The hypothetical chromosome with therapeutic properties can be constructed by the inserting these sequences into HAC at appropriate restriction sites and can then be transfected into Haematopoietic stem cells which can differentiate into CD4+ T cells (Staal et al. 1998) and get expressed. Figure 3 represents a model of such an engineered HAC.
Overcoming Gene Therapy Barriers Using Engineered HAC
Since the therapeutic chromosome is to be delivered using Haematopoietic stem cells as the carrier, immune response/ rejection to this vector is unlikely. The engineered HAC can carry large amounts of therapeutic genes (greater than 100kb), while avoiding insertional inactivation of other genes. It can also be used for other multi-gene disorders by formulating an appropriate gene construct (Chromos Molecular Systems 2005).
Antisense RNA Technology
Figure 4. Regular Transcription and Translation.
Figure 5. Antisense RNA Mechanism.
Antisense RNA is defined as the sequence that is complementary to a functional mRNA sequence. Hence expression of gene of interest' can be regulated by the manipulation of these messenger RNAs.
Figure 6. Action of Dicer.
Messenger RNA (mRNA) is single-stranded. Its sequence of nucleotides is called "sense" because it results in a gene product (protein). Normally, its unpaired nucleotides are read by transfer RNA anticodons as the ribosome proceeds to translate the message. Figure 4 shows the regular transcriptional and translational processes. The principle behind antisense RNA technique is that an antisense nucleic acid sequence base pairs with its complementary sense RNA strand and prevents it from being translated into a protein. Figure 5 shows the formation of RNA duplex. If the cell finds molecules of double-stranded RNA (dsRNA) in the cytoplasm, it uses an enzyme called Dicer' to cut them into fragments containing 19 base pairs with two additional nucleotides at the opposite end of each strand. This interferes with the ability of the mRNA to be translated into a polypeptide by destroying the mRNA when the base-pairing is exact or Repressing its translation when the base-pairing is simply a close match (Figure 6). There is also evidence that antisense strands may sometimes inhibit transcription by binding to complementary sequences on DNA (Arts and Le Grice 1998, Boulmé et al. 1998).
gp120 and its Expression
Glycoprotein 120 (Figure 7) exists as a trimer on the surface of the virion, and binds CD4 on the surface of the target cell, inducing a conformational change in the envelope proteins that in turn allows binding of the virion to a specific subset of chemokine receptors on the cell surface. These receptors normally play a role in chemoattraction, in which Haematopoietic cells move along chemokine gradients to specific sites. The binding of surface gp120, CD4, and the chemokine co receptors produces an additional radical conformational change in gp41. Assembled as a trimer on the virion membrane, this coiled-coil protein springs open, projecting three peptide fusion domains that "harpoon" the lipid bilayer of the target cell. The fusion domains then form hairpin- like structures that draw the virion and cell membranes together to promote fusion, leading to the release of the viral core into the cell interior. gp 120 is encoded by a 1449bp gene fragment (NCBI 2005) and comprises of 481 amino acids, with a molecular weight of about 53922 Daltons. The genetic sequence of gp120 shows a high degree of variation from one strain to another. But, there exists highly conserved regions known as crown regions that exist in its V3 loop region. Residues 307 to 330 have been found to be conserved. The gene construct which is to be Cloned into the HAC is as follows: the gene fragment corresponding to the above mentioned conserved peptide coupled with gene coding for a secretary signal. The expression of this gene construct from engineered HAC will give rise to soluble peptide which will induce both humoral and cell mediated immune responses (Bunnell and Morgan 1998, Pinchuk 2002, Amara et al. 1997, Stanhope et al. 1993, Nor et al. 1998).
Reverse Transcription and Expression of Antisense Gene
HIV-1 Reverse transcriptase is a heterodimer containing 440 amino acid residues. It is involved in the conversion of ssRNA into dsDNA. If this conversion can be blocked by antisense RNA expression, the retroviral life cycle can be inhibited at an early stage itself.
Steps involved in the conversion of are as follows:
1. HIV-1 specific cellular tRNA hybridizes with a complementary region called the primer-binding site (PBS).
2. A DNA segment is extended from tRNA based on the sequence of the retroviral genomic RNA.
3. The viral R and U5 sequences are removed by RNase H.
4. DNA hybridizes with the remaining R sequence at the 3' end.
5. A DNA strand is extended from the 3' end.
6. Most viral RNA is removed by RNase H.
7. A second DNA strand is extended from the viral RNA.
8. Both tRNA and the remaining viral RNA are removed by RNase H.
9. The PBS region of the second strand hybridizes with the PBS region of the first strand.
Figure 7. Protein sequence of gp120. Residues 307-330 are highly conserved in all HIV-1 Isolates.
10. Both DNA strands extend.
The step-by-step process of reverse transcription is represented by Figure 8. The antisense strand of the reverse transcriptase encoding gene, inserted into the therapeutic HAC (1680bp) (NCBI 2005) produces an mRNA that is complementary to the Reverse transcriptase coded by the infecting virus. These complementary mRNA strands bind to each other to form a ds mRNA which is degraded by the enzyme dicer' (Rnase) encoded by the host cell (Rittner and Sczakiel 1991). Therefore, the translation of the viral mRNA is inhibited and prevents the conversion of ssRNA into dsDNA (Berg 2002).
Expression
Figure 8. Step-by-step process involved in Reverse Transcription.
The enzyme Integrase is coded for, by a 564bp long gene fragment containing 288 amino acids (NCBI 2005). Integrase catalyses removal of two bases from the linear end of DNA at the 3' end and gives rise to a 3' recess. Staggered cut 6 nucleotides apart is then made in the genomic DNA and the resulting 5' phosphates are ligated to the 3' OH groups (Figure 9). As explained in the earlier case, the translation of integrase (Parrill 2003) can be blocked by expression of antisense gene fragments which are expressed from the therapeutic chromosome (Rittner and Sczakiel. 1991). Hence, viral integration into host genome can be prevented.
P27 is coded by a 320bp gene fragment. This protein plays a multifunctional role in the infection process. Its interaction with other signaling proteins determines the degree of pathogenicity, also plays a crucial role in viral replication cycle. The expression of anti-sense strand of P27 from the engineered chromosome effectively inhibits its synthesis, thereby preventing the down regulation of cell surface CD4 and Major Histocompatibility class I receptors (MHC - I) (NCBI 2005).
The engineered chromosome can be transferred into the stem cells obtained from the patient or a donor by micromanipulation (Borowski et al. 2000) or lipofection (Felgner et al. 1987, de Jong et al. 2001).
Bone Marrow Transplantation
Figure 9. Mechanism of integration.
Bone marrow Transplantation can be considered to be a suitable method of administering engineered stem cells into the patient, as it is Haematopoietic. For a successful bone marrow transplant, the tissue type of the donor and patient must match very closely, to avoid immune rejection. Hence, the transplant can preferably be from the patient itself (Anthony Nolan Trust 2005). The engineered stem cells can be cultured in vitro (Sigma-Aldrich 2005) and then be infused in vivo into patients bone marrow by the hick man line (Anthony Nolan Trust 2005). After transplantation these stem cells in the bone marrow differentiate into T-cells and express the therapeutic genes.
The expression of the therapeutic chromosome results in the decrease in the intensity of infection.
Conclusion
Bone marrow, being the origin of Haematopoietic cells can be considered to be the ideal site for delivering the engineered stem cells (Chromos Molecular Systems 2005), The Eukaryotic vector (HAC) is autonomously replicating, stably inherited, and contain genetic elements for maximum gene expression (Telenius et al. 1999). The expression of the inserted therapeutic gene sequences (gp120, antisense RT, integrase, p27) in HAC will interfere with various stages of the retroviral life cycle, and hence could be a permanent treatment option for HIV Infection.
Acknowledgements
We wish to sincerely thank our beloved families and friends for their moral support during the drafting process. We are indebted to the Department of Biotechnology at St. Peter's Engineering College, at Anna University, for providing us with constant encouragement during this endeavor.
References
Ali, M et al. (1994) The use of DNA viruses as vectors for gene therapy. Gene Therapy 1(6): 367 - 384.
Amara, A et al. (1997) HIV co receptor down regulation as antiviral principle: SDF-1alpha-dependent internalization of the chemokine receptor CXCR4 contributes to inhibition of HIV replication. Journal of Experimental Medicine 186: 139 - 146.
Anthony Nolan Trust (2005) Bone Marrow Transplants.
Arts, EJ and SFJ Le Grice (1998) Interaction of retroviral reverse transcriptase with template-primer duplexes during replication. Progress in Nucleic Acid Research and Molecular Biology 58: 335 - 394.
Berg, B (2002) Antisense RNA: A Mechanism to Inhibit Gene Expression. Fred Hutchinson Research Center.
Borowski, A et al. (2000) Generation of transgenic mice and germline transmission of a mammalian artificial chromosome introduced into embryos by pronuclear microinjection. Chromosome Research 8: 3 7.
Boulmé, F et al. (1998) Modified (PNA, 2'-O-methyl and phosphoramidate) anti-TAR antisense oligonucleotides as strong and specific inhibitors of in vitro HIV-1 reverse transcription. Nucleic Acids Research 26: 5492 - 5500.
Brenner, MK (1995) Human gene therapy: Progress and problems. Journal of Internal Medicine 237: 229 - 239.
Bunnell, BA and RA Morgan (1998) Gene Therapy for Infectious Diseases. Clinical Microbiology Reviews 11: 42 - 56.
Chromos Molecular Systems (2005) Tech Brief.
Common Wealth Scientific and Industrial Research (2005) Chromosome Engineering and Nanomolecular Biology.
de Jong, G et al. (2001) In-Vitro transfer of a 60 Mb mammalian artificial chromosome into primary cell lines using cationic lipids and a dendrimer. Molecular Therapy 3(5): 194 195.
Fauci, AS (1988) The Human Immuno Deficiency Virus infectivity and mechanisms of Pathogenesis. Science 239: 617.
Felgner, PL et al. (1987) Lipofection: a highly efficient lipid-mediated DNA-transfection procedure. Proceedings of the National Academy of Sciences 84: 7413 7417.
Harrington, JJ et al. (1997) Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nature Genetics 15: 345 - 355
Henning, KA et al. (1999) Human artificial chromosomes generated by modification of a yeast artificial chromosome containing both human alpha satellite and single-copy DNA sequences. Proceedings of the National Academy of Sciences 96(2): 592 - 597
Ikeno M, et al. (1998) Construction of YAC-based mammalian artificial chromosomes. National Biotechnology 16(5): 431 - 439.
McNicholl, I (2005) Side Effects of Antiretroviral Drugs: Related Resources. HIV InSite.
Moyle, G (2001) Mitochondrial toxicity hypothesis for lipoatrophy: a refutation. AIDS 15(3): 413 - 415.
Mulligan, K et al. (2000) Hyperlipidemia and insulin resistance are induced by protease inhibitors independent of changes in body composition in patients with HIV infection. Journal of Acquired Immuno Deficiency Syndrome 23: 35 - 43.
NCBI (2005) Human Immuno Deficiency Virus-1, Complete Genome.
Nor, RI et al. (1998) Immunological and virological analyses of persons infected by human immunodeficiency virus type 1 while participating in trials of recombinant gp120 subunit vaccines. Journal of Virology 72: 1552 - 1576.
Parrill, AL (2003) HIV-1 integrase inhibition: binding sites, structure activity relationships and future perspectives. Current Medical Chemistry 10(18): 1811 - 1824.
Pinchuk, G (2002) Schaum's Outlines Immunology. Immunity and immune system. McGrawHill, New York.
Riethman, HC et al. (2001) Integration of telomere sequences with the draft human genome sequence. Nature 409: 948 951.
Rittner, K and J Sczakiel (1991) Identification and analysis of antisense RNA target regions of the human immunodeficiency virus type 1. Nucleic Acids Research 19: 1421 - 1426.
Schueler MG, et al. (2001) Genomic and genetic definition of a functional human centromere. Science 294: 109 115.
Sigma-Aldrich (2005) Stemline Hematopoietic Stem Cell Expansion Medium.
Staal, FJ et al. (1998) Development of retrovirally marked human T progenitor cells into mature thymocytes. International Immunology 7: 1301 - 1309.
Stanhope, PE et al. (1993) An HIV-1 envelope protein vaccine elicits a functionally complex human CD4+ T cell response that includes cytotoxic T lymphocytes. Journal of Immunology 150: 4672 - 4686.
Stephenson, J (1998) Anti-AIDS Gene Therapy. Journal of the American Medical Association 279: 494.
Telenius, H et al. (1999) Stability of a Functional Murine Satellite DNA-Based Artificial Chromosome across Mammalian Species. Chromosome Research 7: 3 - 7.