Authors: Denali H. Davis, Ty C. Stewart, Sudipta Majumdar, and Avijita Jain
Institution Information: Department of Chemistry, 975 Oakland Avenue, Indiana University of Pennsylvania, Indiana, PA 15705
ABSTRACT
Photodynamic therapy (PDT) is a type of treatment that uses light along with photoactive compounds to treat cancer. Light-activated metal complexes with reduced side effects and increased efficacy are promising anticancer agents for PDT. In this study, the amplification of a 1228 base pair region of plasmid pUC18 DNA was used to assess the impact of the metal complex, [Ru(biq)2(dpp)]2+ (biq = 2,2'-biquinoline, dpp = 2,3-bis(2-pyridyl)pyrazine), on the DNA replication needed for cancer cell proliferation. The metal complex [Ru(biq)_2(dpp)]^2+ undergoes ligand exchange and binds with DNA upon light activation (λ_irr ≥ 550 nm). Herein, we report that the complex [Ru(biq)_2(dpp)]^2+inhibits DNA amplification of pUC18 DNA upon activation with light under hypoxic conditions, as evaluated by Polymerase Chain Reaction (PCR). Upon addition of increasing concentration of the complex, increased inhibition of DNA amplification was observed when irradiated for 30 minutes with visible light (λ_irr ≥ 550 nm). The ability of the complex to inhibit DNA amplification was compared with that of a well-known anticancer drug, cisplatin. This unique property of the complex to bind with DNA under hypoxic conditions and inhibit DNA amplification upon light activation makes this a molecule of interest for PDT.
INTRODUCTION
Preceded by only heart disease, cancer is the second leading cause of death in the United States according to the American Cancer Society. As such, there is a constant search for new anticancer agents with increased efficiency and reduced harmful side effects compared to existing options. Cisplatin, cis-diamminedichloroplatinum(II), is a common chemotherapy drug, currently being used to treat a variety of cancers such as testicular, breast, lung, and bladder cancer. The drug works by binding to DNA, forming covalent cross-links that distort the helical structure and prevent repair and replication, which results in apoptosis (Rosenberg et al., 1965; Rosenberg et al., 1969; Sherman and Lippard, 1987; Wong and Giandomenico, 1999; Zang and Lippard, 2003). Cisplatin has serious side-effects, including nephrotoxicity, neurotoxicity, ototoxicity, and myelosuppression (Hambley, 2001; Herman et al., 2008). Due to these severe side-effects, significant efforts have been made towards discovering safer and more effective alternatives.
In recent years, ruthenium-based complexes have emerged as promising photodynamic therapy (PDT) agents. Ruthenium-based complexes display interesting photophysical and photochemical properties and play an important role in energy and electron transfer processes (Balzani, 2003; Balzani et al., 2008; Balzani et al., 1996, Juris et al., 1988). These complexes have been shown to photocleave DNA via singlet oxygen (1O2) generation (Chouai et al., 2005; Friedman et al., 1990; Grover and Thorp, 1991; Jain et al., 2008; Neyhart et al., 1993). The Metal to Ligand Charge Transfer (3MLCT) state of these complexes undergoes energy transfer to molecular oxygen (3O2) to generate 1O2, which reacts with DNA, cleaving the backbone (Chouai et al., 2005; Friedman et al., 1990; Grover and Thorp, 1991; Jain et al., 2008; Neyhart et al., 1993).
Tumors often have low levels of oxygen; therefore, it is necessary to develop PDT agents that operate in the absence of oxygen. Ru(II) polypyridyl complexes with sterically bulky ligands are known as photocisplatin analogs and have been reported to covalently bind with DNA upon irradiation with visible light in the absence of oxygen (Dickerson et al., 2014; Dmytro et al., 2017; Glazer, 2013; Howerton et al., 2012; Kohler et al., 2017; Wachter et al., 2012; Wyland et al., 2017; Albani et al., 2015; Knoll et al., 2014). The bulky ligands lower the energy of the ligand field (3LF) state relative to the 3MLCT state, thus resulting in photoejection of the ligand (Allen et al., 1984; Caspar and Meyer, 1983; Ford, 1970; Ford, 1982; Garner et al., 2011; Malouf and Ford, 1974). The sterically-strained Ru(II) complexes, [Ru(biq)_2(phen)]^2+ and [Ru(biq)(phen)2]^2+, (biq = 2,2'-biquinoline, phen = 1,10-phenanthroline), undergo photoinduced ligand exchange and bind with DNA under hypoxic conditions after excitation by red light (λ_irr > 600 nm or 650 nm) (Albani et al. 2015). We have recently reported that the Ru(II)Pt(II) bimetallic complex, [Ru(biq)2(dpp)PtCl2]2+ , (dpp = 2,3-bis(2-pyridyl)pyrazine), displays multifunctional covalent binding with DNA upon light activation (λirr ≥ 590nm) through: 1) the [Ptdpp(L)_2]^2+ subunit and 2) open coordination sites of the Ru-based chromophore, [Ru(biq)_2(L)2]^2+ (L = solvent) in the absence of oxygen (Jain et al., 2018).
Polymerase Chain Reaction (PCR) allows for rapid amplification of a DNA molecule and is applicable in various biomedical fields including DNA fingerprinting, detection of bacteria or viruses, and diagnosis of genetic disorders. The efficiency of PCR depends upon structural integrity of a DNA molecule; therefore, the distortion of DNA structure decreases the efficiency of a PCR reaction and the subsequent DNA amplification produced in the PCR (Bingham et al., 1996; Kalinowski et al., 1992; McMahon et al., 1987). Jennerwein and colleagues (1991) have used PCR to quantify DNA lesions produced by the anticancer drug cisplatin with an inhibition sensitivity proportional to the size of amplified DNA (Jennerwein et al., 1991). The complex, [Rh2(O2CCH3)4] has been shown to inhibit the amplification of a 148 base pairs (bp) DNA fragment at a bp/mc (metal complex) ratio of 1:50 when incubated for 24 hours (Rahman et al., 2007). Brewer and coworkers (2013) have demonstrated that the complex [(bpy)2Os(dpp)RhCl2(phen)]3+ (bpy = 2,2´-dipyridyl) acts as a red light-activated agent and inhibits DNA replication and amplification of 670bp DNA at a bp/mc ratio of 50:1 when irradiated with red light for 240 minutes (Wang et al., 2013). Complexes with Iridium centers containing polypyridyl-pyrazine-based ligands have been reported to intercalate into DNA and inhibit DNA amplification (Chandra et al., 2016). Recently, a Zn(II)-based polypyridyl complex has been reported to act as a roadblock for DNA polymerase both in vivo and in vitro (Kumar, 2017).
Figure 1. The structure of complex, [Ru(biq)2(dpp)](PF6)2 (biq = 2,2'-biquinoline, dpp = 2,3-bis(2-pyridyl)pyrazine).
The metal complex under investigation, [Ru(biq)_2(dpp)]^2+, displays intense ligand-based π → π* transitions in the UV region and MLCT transitions in the visible region (Wyland et al., 2017). The complex shows photoactivated exchange of the dpp ligand with the solvent when irradiated with visible light (λirr ≥550 nm) to generate the [Ru(biq)_2(L)_2](PF_6)_2 complex (L = acetonitrile or water). We have recently reported that the [Ru(biq)_2(dpp)]^2+complex displays covalent binding with DNA similar to cisplatin upon photoirradiation with visible light (Wyland et al., 2017).
In this study, the impact of the complex [Ru(biq)_2(dpp)]^2+ on DNA amplification was studied both following incubation in darkness and after being subjected to photolysis using PCR (Figure 1). The biological activity of the complex, [Ru(biq)_2(dpp)]^2+, was compared with that of cisplatin. To the best of our knowledge, the [Ru(biq)_2(dpp)]^2+ complex represents the second example in the literature to display photoinduced inhibition of DNA amplification.
MATERIALS AND METHODS
Reagents and Apparatus
All reagents were used as received unless otherwise noted. Plasmid (pUC18) DNA was purchased from Bayou Biolabs (Metairie, LA). The plasmid DNA was used for the studies because of its simplicity in preparation and low cost. The Lambda DNA/HindIII molecular marker was obtained from Promega (Madison, WI). Electrophoresis grade boric acid, agarose, tris(hydroxymethyl)aminomethane (TRIS), Taq DNA polymerase, and deoxyribonucleotide triphosphates (dNTPs) were all obtained from Fisher Scientific (Waltham, MA). Forward and Reverse primers were obtained from Integrated DNA Technologies (Coralville, IA). The metal complex, [Ru(biq)2(dpp)](PF6)2, was synthesized as previously reported (Wyland et al., 2017).
For light activation, samples were irradiated with light from a 450 W xenon arc lamp equipped with a water infrared (IR) filter (Newport Photonics, Irvine CA) and a 550 nm cutoff filter (Newport Photonics, Irvine CA). Electrophoresis was performed using a Fisher Scientific (Waltham, MA) Model FB300 electrophoresis stage at 150 V (∼35 mA) for 45 minutes. Gels were stained with 0.5 μg/mL ethidium bromide for 30 minutes followed by 30 minutes of destaining in ddH2O. Gels were visualized on a Fisher (Waltham, MA) Biotech UV transilluminator and photographic records were taken using a digital camera equipped with an ethidium bromide filter.
Polymerase Chain Reaction
Each 50 mL PCR sample was prepared with 1.0 mL template containing 5.0 ng supercoiled pUC18 plasmid DNA (alone or mixed with metal complex and either incubated in darkness or subjected to photolysis), 1.0 mL of each 10 mM primer (5’-GGTGAAAAAAAAAAAAAAAAACCTCTGACACATGCAGCTCC -3’ and 5’-CCTCGCTCTGCTAATCCTGTTACCAGTGG -3’), 5.0 mL 10X Taq reaction buffer, 1 mL of Taq DNA polymerase, 1.0 mL 10 mM dNTPs, and 40.0 mL deionized H2O in a 0.5 mL PCR tube. The PCR samples were incubated for 60 minutes prior to being set to run 30 thermal cycles of incubation at 95°C, 70°C, and 68°C over the course of 90 minutes using a BIO-RAD (Hercules, CA) T-100 Thermal Cycler. The PCR products were analyzed by 0.8% agarose gel electrophoresis.
RESULTS
Figure 2. (A) Agarose gel electrophoresis results of PCR amplified products, when DNA was incubated for 60 mins with either cisplatin in dark or with [Ru(biq)2(dpp)]2+under dark condition and after photoirradiation. Lane 1 is the molecular weight marker (24kbp, 9.4kbp, 6.6kbp, 4.4kbp, 2.2kbp), Lane 2 is a positive PCR control showing 1228 bp fragment produced using pUC18 DNA as a template, Lane 3 is the product of PCR using pUC18 DNA incubated with cisplatin at bp/mc ratio of 1:5, Lane 4 is the product of PCR using pUC18 incubated with [Ru(biq)2(dpp)]2+ at bp/mc ratio of 1:5 in the dark. Lane 5 is the product of PCR using the same sample irradiated with visible light (λirr≥ 550 nm) for 30 minutes (bp = base pair, mc = metal complex). (B) The relative band intensity of each PCR product.
The PCR reactions were performed after plasmid pUC18 DNA was incubated for 60 minutes under dark conditions with either cisplatin or the [Ru(biq)_2(dpp)]^2+ complex following photoirradiation. The intense dark band in Lane 2 (Figure 2) shows that a large amount of DNA amplification occurred during the PCR experiment. Incubation of DNA with cisplatin at a ratio of 1:5 (bp/mc) inhibited DNA amplification completely (Lane 3, Figure 2). The DNA amplification was not altered significantly in the presence of the same concentration of the metal complex (bp/mc, 1:5) incubated in dark conditions (Lane 4, Figure 2). In contrast, DNA amplification was completely inhibited when the PCR was conducted using the same concentration of metal complex (bp/mc,1:5) irradiated with visible light (λ_irr ≥ 550 nm) for 30 minutes under argon atmosphere (Lane 5, Figure 2).
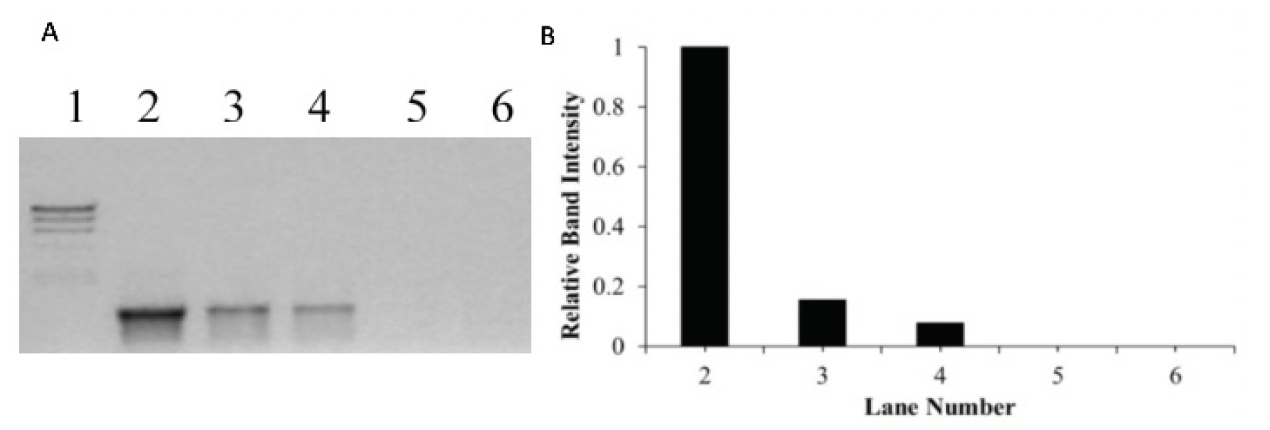
Figure 3. (A) Agarose gel electrophoresis results of PCR amplified product when DNA was incubated for 60 mins with increasing concentrations of metal complex, [Ru(biq)2(dpp)]2+, photolyzed with visible light (visible light at λ_irr ≥ 550 nm for 30 minutes). In Gel A, Lane 1 is the molecular weight marker (24kbp, 9.4kbp, 6.6kbp, 4.4kbp, 2.2kbp), Lane 2 is a positive PCR control showing the 1228 bp fragment produced using pUC18 DNA as a template, Lane 3-6 is the product of PCR using pUC18 incubated with the complex, [Ru(biq)2(dpp)]2+, at bp/mc ratio of 1:1, 1:3, 1:5 and 1:6, respectively (bp = base pair, mc = metal complex). (B) The relative band intensity of each PCR product.
To determine the minimum bp/mc ratio needed for complete inhibition of DNA amplification, PCR was performed in the presence of increasing concentration of the [Ru(biq)2(dpp)]2+ complex, irradiated with visible light (λ_irr ≥ 550 nm) for 30 minutes. A decrease in the formation of PCR product was observed with an increase in concentration of the [Ru(biq)_2(dpp)]^2+ complex when irradiated with visible light (λ_irr ≥ 550 nm) under hypoxic conditions (Lanes 3-6, Figure 3). Complete inhibition of DNA amplification was observed at bp/mc ratio of 1:5 (Figure 3A and B).
Figure 4. (A) Agarose gel electrophoresis on PCR amplified product when DNA was incubated for 60 mins with increasing concentrations of cisplatin. Lane 1 is the molecular weight marker (24 kbp, 9.4 kbp, 6.6 kbp, 4.4 kbp, 2.2 kbp), Lane 2 is a positive PCR control showing the 1228 bp fragment produced using pUC18 DNA as a template, Lane 3-6 is the product of PCR using pUC18 incubated with cisplatin, at bp/cisplatin ratio of 1:1, 1:3, 1:5 and 1:6, respectively (bp = base pair, mc = metal complex). (B) The relative band intensity of each PCR product.
The impact of similar concentrations of cisplatin on DNA amplification was studied by incubating DNA with increasing concentrations of cisplatin. When DNA was incubated with cisplatin in the dark at a bp/cisplatin ratio of 1:1, 1:3, 1:5 and 1:6, complete inhibition of DNA amplification was observed (Figure 4).
DISCUSSION
These results show that the complex [Ru(biq)_2(dpp)]^2+ inhibits DNA amplification upon light activation though an oxygen-independent mechanism, indicating its potential as a PDT agent. Brewer and coworkers (2013) have observed similar inhibition of DNA amplification upon light activation when DNA was incubated with the [(bpy)_2Os(dpp)RhCl_2(phen)]^3+ complex (Wang et al., 2013).
To confirm that the decrease in the formation of the PCR product is due to the interaction of metal complex with DNA upon light activation and not due to the direct inhibition of the Taq polymerase, a similar experiment was performed with fivefold excess of Taq polymerase, keeping the other parameters constant. At this high concentration of Taq polymerase, no PCR product was observed when incubated with the [Ru(biq)_2(dpp)]^2+ complex at a ratio of 1:5 (bp/mc) and irradiated with visible light (λ_irr ≥ 550 nm) for 30 minutes, confirming that the inhibition of DNA amplification is due to the interaction of metal complex with DNA rather than direct inhibition of the Taq polymerase.
In order to determine the minimum concentration of photolyzed metal complex needed for complete inhibition of DNA amplification, PCR reactions were performed where DNA was incubated with increasing concentration of metal complex (bp/mc ratios of 1:1, 1:3, 1:5 and 1:6) and irradiated with visible light (Figure 3). The inhibition of DNA amplification at a higher concentration of the metal complex is due to the binding of the metal complex with DNA upon light activation, which leads to the distortion of DNA structure and therefore, decreases the efficiency of a PCR reaction. Chandra and Kumar (2016, 2017) have shown similar inhibition of DNA amplification due to interactions with iridium and ruthenium-containing compounds in the dark (Chandra et al., 2016; Kumar, 2017). When DNA was incubated with similar concentrations of cisplatin in the dark at bp/cisplatin ratio of 1:1, 1:3, 1:5 and 1:6, complete inhibition of DNA amplification was observed (Figure 4). Cisplatin inhibits DNA amplification by binding to the DNA segment, altering its shape, and thus preventing it from being replicated.
Selective inhibition of DNA amplification upon light activation under hypoxic conditions makes the designed [Ru(biq)_2(dpp)]^2+ complex a molecule of interest for PDT. Additionally, the PCR conditions used in these experiments included 30 cycles of incubation at 95°C, 70°C, and 68°C over the course of 90 minutes, indicating substantial thermal stability of the photomodified DNA product responsible for inhibition of DNA amplification. The unique properties of this complex are imparted by lower energy of the ^3LF state relative to the ^3MLCT state, thus resulting in ejection of the dpp ligand upon photoactivation and creating open coordination sites for covalent binding with DNA. As a result of covalent interactions with DNA upon photoirradiation, the [Ru(biq)_2(dpp)]^2+ complex acts as a roadblock for DNA polymerase.
These studies show that the complex [Ru(biq)_2(dpp)]^2+ inhibits DNA amplification upon light activation, demonstrating promise as a potential PDT agent. This complex undergoes ligand dissociation upon activation with light, thereby opening the coordination sites to bind with DNA. The mechanism of DNA damage does not require reactive oxygen species. Upon light activation at a 1:5 bp/mc ratio, the DNA damage caused by the complex is sufficient to inhibit DNA amplification by PCR. Furthermore, this modification to DNA is stable at the 95°C thermal cycling needed for PCR. Future work is underway to investigate the in vivo bioreactivity of [Ru(biq)2(dpp)]2+ and related metal complexes using cancer cell lines.
ACKNOWLEDGMENTS
This work was supported by the Indiana University of Pennsylvania Senate Award (4012310571).
REFERENCES
Albani, B. A.; T., W.; Durr, C. B.; Turro, C. (2015) Steric and electronic factors associated with the photoinduced ligand exchange of bidentate ligands coordinated to Ru(II). Photochemistry and Photobiology, 91, 616-623, available: 10.1111/php.12392
Allen, G. H.; White, R. P.; Rillema, D. P.; Meyer, T. J. (1984) Synthetic control of excited-state properties. Tris-chelate complexes containing the ligands 2,2'-bipyrazine, 2,2'-bipyridine, and 2,2'-bipyrimidine. Journal of the American Chemical Society, 106 (9), 2613-2620, available: https://doi.org/10.1021/ja00321a020
Balzani, V. (2003) Photochemical molecular devices. Photochemical and Photobiological Sciences, 2, 459-476, available: https://doi.org/10.1039/B300075N
Balzani, V., Juris, A., and Venturi M. (1996) Luminescent and redox active polynuclear transition metal complexes. Chemical Reviews, 96, 759-833, available: https://doi.org/10.1021/cr941154y
Balzani, V.; Bergamini, G.; Ceroni, P. (2008) From the photochemistry of coordination compounds to light-powered nanoscale devices and machines. Coordination Chemistry Reviews, 252 (23–24), 2456-2469, available: https://doi.org/10.1016/j.ccr.2007.11.009
Bingham, J. P.; Hartley, J. A.; Souhami, R. L.; Grimaldi, K. A. (1996) Strand-Specific Measurement of Cisplatin-Induced DNA Damage and Repair Using Quantitative PCR. Nucleic Acids Research, 24 (5), 987-989, available: 10.1093/nar/24.5.987
Caspar, J. V.; Meyer, T. J. (1983) Photochemistry of MLCT excited states. Effect of nonchromophoric ligand variations on photophysical properties in the series cis-Ru(bpy)2L22+. Inorganic Chemistry, 22 (17), 2444-2453, available: https://doi.org/10.1021/ic00159a021
Chandra, F.; Kumar, P.; Tripathi, S. K.; Patra, S.; Koner, A. L. (2016) Iridium Complexes as a Roadblock for DNA Polymerase during Amplification. ChemMedChem, 11 (13), 1410-1414, available: https://doi.org/10.1002/cmdc.201600101
Chouai, A.; Wicke, S. E.; Turro, C.; Bacsa, J.; Dunbar, K. R.; Wang, D.; Thummel, R. P. (2005), Ruthenium(II) Complexes of 1,12-Diazaperylene and Their Interactions with DNA. Inorganic Chemistry, 44 (17), 5996-6003, available: https://doi.org/10.1021/ic0485965
Dickerson, M.; Sun, Y.; Howerton, B.; Glazer, E. C.(2014) Modifying Charge and Hydrophilicity of Simple Ru(II) Polypyridyl Complexes Radically Alters Biological Activities: Old Complexes, Surprising New Tricks. Inorganic Chemistry, 53 (19), 10370-10377, available: https://doi.org/10.1021/ic5013796
Dmytro, H.; K., H. D.; Leona, N.; Sean, P.; C., G. E. (2017) Photochemical Properties and Structure–Activity Relationships of RuII Complexes with Pyridylbenzazole Ligands as Promising Anticancer Agents. European Journal of Inorganic Chemistry, 2017 (12), 1687-1694, available: https://doi.org/10.1002/ejic.201601450
Ford, P. C. (1970) Properties and reactions of ruthenium (II) amine complexes. Coordination Chemistry Reviews, 5, 75-99, available: https://doi.org/10.1016/S0010-8545(00)80075-2
Ford, P. C. (1982) The ligand field photosubstitution reactions of d6 hexacoordinate metal complexes. Coordination Chemistry Reviews, 44, 61-82, available: https://doi.org/10.1016/S0010-8545(00)80517-2
Friedman, A. E.; Chambron, J. C.; Sauvage, J. P.; Turro, N. J.; Barton, J. K. (1990) A molecular light switch for DNA: Ru(bpy)2(dppz)2+. Journal of the American Chemical Society, 112 (12), 4960-4962, available: https://doi.org/10.1021/ja00168a052
Garner, R. N.; Joyce, L. E.; Turro, C. (2011) Effect of Electronic Structure on the Photoinduced Ligand Exchange of Ru(II) Polypyridine Complexes. Inorganic Chemistry, 50 (10), 4384-4391, available: https://doi.org/10.1021/ic102482c
Glazer, E. C. (2013) Light-activated metal complexes that covalently modify DNA. Israel Journal of Chemistry, 53, 391-400, available: https://doi.org/10.1002/ijch.201300019
Grover, N.; Thorp, H. H. (1991) Efficient electrocatalytic and stoichiometric oxidative cleavage of DNA by oxoruthenium(IV). Journal of the American Chemical Society, 113 (18), 7030-7031, available: https://doi.org/10.1021/ja00018a048
Hambley, T. (2001) Platinum binding to DNA: structural controls and consequences. Dalton Transactions, 2711-2718, available: https://doi.org/10.1039/B105406F
Herman, A.; Tanski, J. M.; Tibbetts, M. F.; Anderson, C. M. (2008) Synthesis, Characterization and in vitro Evaluation of a Potentially Selective Anticancer, mixed-metal Ruthenium(III)-Platinum(II) trinuclear complex. Inorganic Chemistry, 47, 274-280, available: https://doi.org/10.1021/ic062419h
Howerton, B. S.; Heidary, D. K.; Glazer, E. C. (2012) Strained Ruthenium Complexes Are Potent Light-Activated Anticancer Agents. Journal of American Chemical Society, 134 (20), 8324-8327, available: https://doi.org/10.1021/ja3009677
Jain, A.; Slebodnick, C.; Winkel, B. S. J.; Brewer, K. J. (2008) Enhanced DNA photocleavage properties of Ru(II) terpyridine complexes upon incorporation of methylphenyl substituted terpyridine and/or the polyazine bridging ligand dpp (2,3-bis(2-pyridyl)pyrazine). Journal of Inorganic Biochemistry, 102 (10), 1854-1861, available: 10.1016/j.jinorgbio.2008.06.004
Jain, A.; Wyland, K. R.; Davis, D. H. (2018) Redox, spectroscopic, photo-induced ligand exchange, and DNA interaction studies of a new Ru(II)Pt(II) bimetallic complex. Journal of Coordination Chemistry, 71 (2), 231-242, available: https://doi.org/10.1080/00958972.2017.1420788
Jennerwein, M. M.; Eastman, A. (1991) A polymerase chain reaction-based method to detect cisplatin adducts in specific genes. Nucleic Acids Research, 19 (22), 6209-6214, available: 10.1093/nar/19.22.6209
Juris, A.; Balzani, V.; Barigelletti, F.; Campagna, S.; Belser, P.; von Zelewsky, A. (1988) Ru(II) polypyridine complexes: photophysics, photochemistry, electrochemistry, and chemiluminescence. Coordination Chemistry Reviews, 84, 85-277, available: https://doi.org/10.1016/0010-8545(88)80032-8
Kalinowski, D. P.; Illenye, S.; Vanhouten, B. (1992) Analysis of DNA damage and repair in murine leukemia-L1210 cells using a quantitative polymerase chain reaction assay. Nucleic Acids Research, 20 (13), 3485-3494, available: 10.1093/nar/20.13.3485
Knoll, J. D.; Albani, B. A.; Durr, C. B.; Turro, C. (2014) Unusually Efficient Pyridine Photodissociation from Ru(II) Complexes with Sterically Bulky Bidentate Ancillary Ligands. Journal of Physical Chemistry A, 118 (45), 10603-10610, available: https://doi.org/10.1021/jp5057732
Kohler, L.; Nease, L.; Vo, P.; Garofolo, J.; Heidary, D. K.; Thummel, R. P.; Glazer, E. C. (2017) Photochemical and Photobiological Activity of Ru(II) Homoleptic and Heteroleptic Complexes Containing Methylated Bipyridyl-type Ligands. Inorganic Chemistry, 56 (20), 12214-12223, available: https://doi.org/10.1021/acs.inorgchem.7b01642
Kumar, A. (2017) A small-molecule acts as a ‘roadblock’ on DNA, hampering its fundamental processes. Journal of Inorganic Biochemistry, 176, 134-139, available: https://doi.org/10.1016/j.jinorgbio.2017.08.023
Malouf, G.; Ford, P. C. (1974) Photochemical reaction pathways of ruthenium(II) complexes. Evidence regarding the reactive excited state(s) from metal-to-ligand charge transfer excitation of pentaamine(pyridine)ruthenium(2+) and related complexes. Journal of the American Chemical Society, 96 (2), 601-603, available: https://doi.org/10.1021/ja00809a056
McMahon, G.; Davis, E.; Wogan, G. N. (1987) Characterization of c-Ki-ras oncogene alleles by direct sequencing of enzymatically amplified DNA from carcinogen-induced tumors. Proceedings of the National Academy of Sciences of the United States of America, 84 (14), 4974-4978, available: 10.1073/pnas.84.14.4974
Neyhart, G. A.; Grover, N.; Smith, S. R.; Kalsbeck, W. A.; Fairley, T. A.; Cory, M.; Thorp, H. H. (1993) Binding and kinetics studies of oxidation of DNA by oxoruthenium(IV). Journal of the American Chemical Society, 115 (11), 4423-4428, available: https://doi.org/10.1021/ja00064a001
Rahman, M. M.; Yasuda, H.; Katsura, S.; Mizuno, A. (2007) Inhibition of endonuclease cleavage and DNA replication of E. coli plasmid by the antitumor rhodium(II) complex. Archives of Biochemistry and Biophysics, 464 (1), 28-35, available: 10.1016/j.abb.2007.03.041
Rosenberg, B.; Camp, V.; Kriagas, T. (1965) Inhibition of Cell Division in Escherichia coli by Electrolysis Products from a Platinum Electrode. Nature, 205, 698-699, available: https://doi.org/10.1038/205698a0
Rosenberg, B.; L, V. C.; Trosko, J. E.; Mansour, V. H. (1969) Platinum Compounds: a New Class of Potent Antitumour Agents. Nature, 222, 385-387, available: https://doi.org/10.1038/222385a0
Sherman, S. E.; Lippard, S. J. (1987) Structural aspects of platinum anticancer drug interactions with DNA. Chemical Reviews, 87, 1153-1181, available: https://doi.org/10.1021/cr00081a013
Wachter, E.; Heidary, D. K.; Howerton, B. S.; Parkin, S.; Glazer, E. C. (2012) Light-activated ruthenium complexes photobind DNA and are cytotoxic in the photodynamic therapy window. Chemical Communications, 48, 9649-9651, available: https://doi.org/10.1039/C2CC33359G
Wang, J.; Newman, J., Jr.; Higgins, S. L.; Brewer, K. M.; Winkel, B. S.; Brewer, K. J. (2013) Red-light-induced inhibition of DNA replication and amplification by PCR with an Os/Rh supramolecule. Angewandte Chemie (International ed. in English), 52 (4), 1262-1265, available: https://doi.org/10.1002/anie.201207083
Wong, E.; Giandomenico, C. M. (1999) Current Status of Platinum-Based Antitumor Drugs. Chemical Reviews, 99, 2451-2466, available: https://doi.org/10.1021/cr980420v
Wyland, K. R.; Hoffman, E. E.; Jain, A. (2017) DNA interaction studies and photoinduced ligand exchange kinetics of a sterically strained ruthenium(II) complex. Inorganica Chimica Acta, 454, 62-66, available: https://doi.org/10.1016/j.ica.2016.06.008
Zang, C. X.; Lippard, S. (2003) New metal complexes as potential therapeutics. Current Opinion in Chemical Biology, 7, 481-489, available: https://doi.org/10.1016/S1367-5931(03)00081-4


2 (biq = 2,2'-biquinoline, dpp = 2,3-bis(2-pyridyl)pyrazine).](https://images.squarespace-cdn.com/content/v1/5443d7c7e4b06e8b47de9a55/1564604400036-2FGDYW8N609GDI26TYVY/image-asset.png)
![Figure 2. (A) Agarose gel electrophoresis results of PCR amplified products, when DNA was incubated for 60 mins with either cisplatin in dark or with [Ru(biq)2(dpp)]2+under dark condition and after photoirradiation. Lane 1 is the molecular weight ma…](https://images.squarespace-cdn.com/content/v1/5443d7c7e4b06e8b47de9a55/1564604608076-0THE65ARR9I9DGBEY0C3/Screen+Shot+2019-07-31+at+3.19.03+PM.png)
![Figure 3. (A) Agarose gel electrophoresis results of PCR amplified product when DNA was incubated for 60 mins with increasing concentrations of metal complex, [Ru(biq)2(dpp)]2+, photolyzed with visible light (visible light at λ_irr ≥ 550 nm for 30 m…](https://images.squarespace-cdn.com/content/v1/5443d7c7e4b06e8b47de9a55/1564604640623-BTI6FDLTJG8V3YLWU8GM/image-asset.png)