Author: Marnin D. Wolfe
Institution: Department of Biology, Eckerd College, St. Petersburg, FL 33711 USA
Date: March 2007
ABSTRACT
Effective conservation strategies require that agencies account for genetic diversity and its geographic distribution within threatened species. Unionid mussels are among the most endangered taxa in North America, with between 55% and 72% of species having some protected status, and many others declining rapidly. With so few populations of these mussels safe from anthropogenic impacts, it is vital that we study relatively undisturbed species to understand historical genetic distributions. The fat mucket, Lampsilis siliquoidea, was assayed for genetic variation in Darby Creek, a drainage system in central Ohio, where it is a member of a relatively undisturbed bivalve community. Genotypes of mussels from five locations in the basin were determined at four microsatellite loci via polyacrylamide gel electrophoresis. All populations deviated from Hardy-Weinberg expectations and exhibited heterozygote deficiencies. Most (96%) of the total variation occurred within populations and genetic divergence (FST) was not correlated with geographic distance. These results demonstrate significant levels of variation within populations as well as a higher level of differentiation, given the geographic scale, than endangered unionids. The study predicts that small-stream species will be highly distinct among-drainages and therefore require separate management strategies.
INTRODUCTION
There is overwhelming evidence that declines in population sizes lead to reduced genetic diversity through random genetic drift and inbreeding (Avise 2004). Although it is difficult to prove decreased heterozygosity or genetic diversity as cause for decreased fitness, a number of studies have shown significant correlations in a variety of taxa, including: Asiatic lions (Panthera leo persica; O'Brien and Evermann 1988), Sonoran topminnows (Quattro and Vrijenhoek 1989), and gray wolves (Canis lupus) (Wayne et al. 1991). Saccheri et al. (1998) demonstrated the first direct linkage between inbreeding and extinction of subpopulations in a large metapopulation of the Glanville fritillary butterfly (Melitaea cinxia). In a survey of 170 threatened taxa worldwide, Spielman et al. (2004) found that 77% of threatened taxa had significantly lower heterozygosity than their closest nonthreatened relative. The negative effects of decreased genetic variability are the most extreme in the most highly endangered species such as the plains pigeon (Columba inornata wetmorei; Young et al. 1998), the greater prairie chicken (Tympanuchus cupido; Bouzat et al. 1998), and the cheetah (Acinonyx jubatus; O'Brien et al. 1996).
Freshwater mussels, also called clams or naiads, are bivalves belonging to the superfamily Unionoidea and comprise one of the most endangered groups of organisms in North America. Over 70% of unionid taxa are listed as extinct, threatened, or of special concern (Williams et al. 1993). Aquatic bivalves play important roles in both lotic and lentic ecosystems and their absence can have far-reaching and complex consequences (Vaughn and Hakenkamp 2001, Strayer et al. 2004). In order to preserve these important organisms, it is necessary to design conservation strategies that consider both the abundance and genetic diversity of unionids (Berg et al. 1998). The unionids are notable in that their glochidia require incubation in a vertebrate host for survival to adulthood. Many unionids suspend glochidia on threads of mucus, which facilitate infection of a host by either contact with or consumption by the host, while others lure a host to them and pump glochidia through the excurrent siphon, into the host. Glochidia form a protective epithelium on fins, under scales, or on gills of the host fish and grow until capable of self-sufficiency, finally detaching and settling into the substrate (Fuller 1974).
Threats to freshwater bivalves are many and severe. Seven percent of nearly 300 unionid species (Strayer et al. 2004) are listed as possibly extinct in North America (Williams et al. 1993). Many extinctions may have been caused by damming, which is known to adversely impact 30-60% of all native mussels both upstream and downstream. Dams cause shore erosion and siltation, which suffocates bivalves (Watters 1996) and destroy the unionid reproductive cycle through loss of host species (Bogan 1993). Commercial harvest of freshwater mussels has also been severe. Until the advent of plastics, mussel shell was the primary source of buttons. Today the shells are commercially harvested for the production of seed pearls in the pearl culture industry (Strayer et al. 2004; Williams et al. 1993; Bogan 1993). Invasive clams such as Corbicula fluminea, the Asiatic clam, and Dreissena polymorpha, the zebra mussel, are decimating native mussels in several parts of North America because they are capable of much faster rates of filtration and thus out-compete unionids for food and habitat (Williams et al. 1993).
With freshwater mussels undergoing catastrophic declines worldwide, there is a great need for effective conservation strategies. A key component of effective conservation is examination of genetic diversity, which is necessary for sustaining population viability. There are two commonly described components to genetic variation: that within a population and that among separate populations (Berg et al. in press). Within-population variation is primarily affected by genetic drift, the probability of inbreeding, and the maintenance and loss of rare alleles. Meanwhile, variation among populations should reflect the geographic structure of a species' genome as it is affected by local adaptations, selection, reduced or elevated gene flow and isolation (Avise 2004). Thus far, the majority of studies of mussel population genetics have been assays of genetic variation in highly endangered species (Liu et al. 1996; Turner et al. 2000; Geist and Kuehn 2005). Unfortunately, few of these studies focus on mussel species that are common and stable. Genetic structure of highly endangered species tells us little of what normal genetic variation of freshwater mussels might look like in a robust set of populations. Such knowledge is vital as a target for restoration of depleted species. In an effort to quantify genetic variation in one such system, Berg et al. (in press) assayed allozyme variation of the three most common mussel species in the Darby Creek watershed in central Ohio. Darby Creek represents a unique system with an unusually high diversity (38 species) and abundance of mussels. The creek has been largely spared the damages of damming, species invasions and suburban development (EPA Ohio 2002). In fact, in 1991 the Nature Conservancy gave Darby Creek recognition as one of the world's Last Great Places. Thus the system represents an optimal location to study mussel populations. All three species studied (Elliptio dilatata, Ptychobranchus fasciolaris, and Lampsilis siliquoidea) possessed genetic variation both within and among populations greater than most unionids. Significantly higher levels of population differentiation were detected in L. siliquoidea than in either of the other two species, despite a complete lack of geographic correlation between distance and differentiation.
In an attempt to gain further understanding of Darby creek L. siliquoidea population structure, the present study used a highly variable class of molecular marker, microsatellites, to assay the relationship between gene flow and genetic drift at a small scale. It was expected that microsatellite loci of L. siliquoidea maintain similar patterns to allozymes in the Darby watershed, but could provide enough variability to elucidate further patterns in the system, adding robustness to current understanding of this species. The microsatellite data are valuable in a conservation context. The Darby Creek L. siliquoidea data set can be used as a model of genetic variation in a mussel species relatively free of adverse anthropogenic effects, while providing insight into the genetic structure of a small-stream mussel.
MATERIALS AND METHODS
Study Area
This study was conducted in Darby Creek, located in central Ohio. The Darby Creek basin covers approximately 143,744 hectares (555 square miles) predominantly covered by high quality agricultural landscapes in six counties: Champaign, Franklin, Logan, Madison, Pickaway, and Union (Figure 1). The creek drains into the Scioto River, part of the Ohio River basin south of Columbus. There is considerable interest in conservation of the creek system because of its richness of mussel (38) and fish (94) species, exceptionally high numbers for a small mid-western stream (Watters 1996).
Sample Collection
Mussels were sampled at three sites along a 60-km section of Big Darby Creek in Union and Madison Counties and at two sites along 28 kilometers of Little Darby Creek in Madison County. L. siliquoidea (Barnes, 1823) were collected at three sites on Big Darby Creek (river miles 39.4, 62.5, 65.7; measured from confluence with the Scioto River) and two sites on Little Darby Creek (river miles 7.7, 12.0 measured from confluence with Big Darby Creek) in May and July of 1998. Mussels were collected by snorkeling in water < 1.5 meters deep. At each site, 20-50 specimens were collected. Small portions of mantle tissue were excised nondestructively, with the exception of a small number (< 5 per site) of whole organisms taken from each population, and flash-frozen in liquid nitrogen and stored in a -80oC freezer.
Collection of Molecular Data
Eleven of 15 primer sets designed to amplify microsatellite loci in the endangered unionid L. abrupta (Eackles and King 2002) were obtained. Prior to PCR amplification of a sample, DNA was extracted from 30-35mg of mantle tissue using DNEasy Tissue Kits (Qiagen). Whole genomic DNA was then quantified using a NanoDrop 2200 spectrophotometer and diluted to a standard concentration of 15ng/ul. Of the 11 available primers, four (LabC67, LabD31, LabD111, and LabD206) were optimized for use with L. siliquoidea.
article_944_order_0
PCR amplifications consisted of 150ng (200ng for Primer 67) of genomic DNA, 1x PCR buffer (10mM Tris-HCl, pH 8.3, 50mM KCl), 2.5mM (Primer 206 and 31) and 3.0mM (Primer 67 and 111) MgCl2, 0.15% Tween-20, 0.25 mM dNTPs, approximately 0.5 µM forward and reverse primer, and 0.4U Taq DNA polymerase in a total volume of 25 µL. Amplifications were carried out on a PTC-200 DNA Engine Gradient Cycler (MJ Research) using the following conditions: 94°C initial heating for 2 minutes followed by 45 cycles (50 cycles for primer 67) at 94°C for 40s, 40s at annealing temperatures (52°C for primers 67 and 111; 60°C for primers 206 and 31), and extension at 72°C for 1 minute, concluding with a final extension at 72°C for 10 minutes (20 minutes for primer 67).
Amplified products were separated on 8% polyacrylamide gels using TBE buffer at 80V for 2.5 hours. Gels were developed in ethidium bromide baths for 15 minutes. Visualization was done with a FluorChem 8800 Imaging System (Alpha Innotech Corporation) under UV lighting. Alleles were scored using the AlphaImager 3300 program for Windows by the same company.
Analysis
Statistical analyses were conducted using GeneAlEx Genetic Analysis Software (Peakall and Smouse 2005). Descriptive statistics employed included: mean number of alleles per locus, percent polymorphic loci (polymorphic loci are those in which the most common allele has a frequency ≤ 95%) and Wright's F-statistics. Tests for deviation of genotype frequencies from Hardy-Weinberg expectation and population assignment were also conducted. Chi-squared goodness-of-fit tests were used to determine if observed heterozygosity values were significantly different from expected heterozygosity.
Analysis of molecular variance (AMOVA) was used to examine hierarchical partitioning of genetic variation in the system. Differentiation was examined between individuals in a population and among populations within the sampling area on Darby Creek. Population differentiation was determined from FST values calculated via both Wright's method (e.g. FST above; Wright 1931) and the AMOVA technique (described in Peakall and Smouse 2005). Conformance of genotype frequencies with Hardy-Weinberg expectation was examined using the HS and HI values. Additionally, FST values were used to test pairwise population correlation between FST and geographic distance to test gene flow-drift equilibrium (Kimura 1953; Kimura and Weiss 1964). Finally, a population assignment test was performed. Individuals were assigned to a population based on their allelic makeup as compared to the allele frequencies in a given population. Should an individual be assigned to a population other than the one in which it was collected, recent dispersal from the assigned population can be inferred (Waser et al. 2001).
RESULTS
Within-population variation
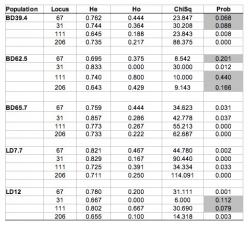
There were a total of 101 individuals remaining in storage from the initial allozyme study that were used in the present study. Four L. abrupta microsatellite loci were successfully amplified from L. siliquoidea: LabC67, LabD31, LabD111, and LabD206. Success rates for microsatellite amplification were moderate to high, as follows: LabC67 was amplified in 83% of individuals, LabD31 in 45%, LabD111 in 67%, and LabD206 in 93%. All four loci were highly polymorphic, as expected with a microsatellite marker (Table 1). LabC67 had 9 alleles, LabD31 had 17, LabD111 had 10, and LabD206 had 9. The number of alleles was negatively correlated with average sample size per locus (r=0.915). However, the sample size (n=4 loci) did not permit significance tests.
article_944_order_4
Mean observed heterozygosity was 0.314 as compared to a mean Hardy-Weinberg expected heterozygosity of 0.745 for an overall deficit of heterozygotes (see Table 2). At the LabC67 locus, all locations deviated significantly except for BD39.4 and BD62.5. LabD31 was significantly different from HW expectations except at BD39.4 and LD12. LabD111 deviated from HW expectation at all sites except BD62.5 and LD12. The LabD206 locus in all populations except BD62.5 deviated significantly from Hardy-Weinberg expectation (P 0.05, n=5 populations) between heterozygosity of allozymes and that of microsatellites. There is little literature on unionid microsatellite variation as yet. Recently, Kelly and Rhymer (2005) reported average H of 0.68 in L. cariosa using microsatellites. Preliminary study of the congener L. abrupta (Eackles and King 2005) found H = 0.469, however sample sizes were small and among-population variation was not considered. The unionoid Margaritifera margaritifera, has an average H of 0.289 for microsatellite markers (Geist and Kuehn 2005). Interestingly, the invasive zebra mussel, Dreissena polymorpha has been shown to possess heterozygosities up to 0.58 (Astanei et al. 2005). Another invertebrate, the marine sponge Crambe crambe had an average heterozygosity of 0.506 (Duran et al. 2004). Thus, there seems to be a wide range of variability in microsatellite heterozygosity among species. Darby populations are within the range demonstrated for invertebrates.
article_944_order_2
L. siliquoidea utilizes centrarchids and minnows as host fishes (Ohio State University Museum of Biological Diversity (MBD) Mussel / Host Database; http://128.146.250.63/Musselhost/). Berg et al. (in press) suggested that the low dispersal ranges of host fishes, which average 6.5 km seasonally but can extend occasionally up to 75 km (Lyons & Kanehl 2002), lead to patchy distributions and poor colonization ability. If host fish dispersal is so low, populations likely breed primarily within themselves, leading to inbreeding which results in low heterozygosity. Indeed, a high value (0.577) was recorded for the inbreeding coefficient (FIS).
The Darby Creek system was also largely heterozygote deficient as compared to Hardy-Weinberg expectations. Indeed, the initial study using allozymes in Darby Creek found significant deficiencies in all three species studied (Berg et al. in press). In the study, Berg et al. attempted to explain these deficiencies in terms of deviation from one or more of the assumptions of Hardy-Weinberg equilibrium. Assortative mating is unreported and indeed very unlikely given unionid mating systems (Fuller 1974) and is therefore unlikely here. No locus is consistently deficient in every population so selection seems unlikely. The Whalund effect, which refers to a lower heterozygosity than expected as a result of significant isolation of populations, is ruled out, as the same common alleles are present in many or all of the populations. L. siliquoidea has not been documented as hermaphroditic; the species is sexually dimorphic (Jass and Glenn 2004), and thus this cannot be a source of homozygote excess.
Mutations of microsatellite markers are frequent enough to produce the high variability that makes them useful to population genetics, but not so frequent as to cause deviation from Hardy-Weinberg expectations. Additionally, mutations of microsatellites are generally believed to be in the form of additions (Balloux and Lugon-Moulin 2002), creating new alleles, which is more likely to account for heterozygote excess. Null alleles are alleles that do not amplify because of mutations at priming sites and are one explanation for at least some deficiency. Especially given that all four loci have non-amplifying individuals (Foltz, 1986). Sample size does not correlate with allelic richness and therefore is not likely accountable for heterozygote deficiency. Berg et al. (in press) suspected that high genetic drift and inbreeding caused the deficiencies in Darby Creek. Low host fish dispersal is therefore the most likely explanation for heterozygote deficiency when compared to Hardy-Weinberg expectations.
The level of among-population variation found in populations of L. siliquoidea in Darby Creek (overall FST = 0.071) was above that found in several studies (Berg et al. 1998; Astanei et al. 2005). AMOVA resulted in a lower value for FST (0.037). However, the two methods were positively correlated suggesting the values vary together. Berg et al. (in press) recorded a higher value of differentiation using allozymes in Darby Creek (0.134). The degree of differentiation in L. siliquiodia in Darby Creek is more than in many, but not all unionids; most notably differentiation is twice that found in more than 2500 km of the Ohio River basin for Quadrula quadrula, FST = 0.031 (Berg et al. 1998). In contrast, L. cariosa was more differentiated (FST = 0.086; Kelly and Rhymer 2005) than L. siliquoidea. M. margaritifera was also more highly differentiated with FST as high as 0.374 among several highly isolated drainages (Geist and Kuehn 2005). Lowest of all, the invasive zebra mussel D. polymorpha ranged in differentiation from FST = -0.1 to 0.04, a result expected of founding populations of invasive organisms (Astanei et al. 2005).
At gene flow-drift equilibrium, genetic distance between populations increases with increasing geographic distance. At greater distance genetic drift is more influential (the amount of drift is independent of the amount of gene flow) relative to gene flow as flow occurs less and less with increasing distance (Kimura 1953). Thus measures of genetic distance such as FST are expected to increase positively with geographic distance (Kimura 1953; Kimura and Weiss 1964).
As seen in Figure 3 there is no apparent geographic pattern to molecular variation and isolation-by-distance is not apparent in Darby Creek. Likewise, the allozyme study found no geographic pattern to variation (Berg et al. in press). Many mussels exhibit isolation by distance at some scale (Berg et al. 1998; Geist and Kuehn 2005) and this is expected given the linear nature of river drainages and unionid dependence on the vagility of host fishes. So if the population is not exhibiting the equilibrium conditions of the model they are thought to adhere to, then what is going on between gene flow and drift to create these conditions?
Hutchison and Templeton (1999) developed a method for identifying gene-flow drift equilibrium. Deviation from equilibrium occurs when a region has been colonized too recently for genetic drift to occur, or when the degree of dispersal is inhibited within the region or parts of the region causing uneven or decreased gene flow. There are four cases distinguishing equilibrium and non-equilibrium states. Case one is described as "regional equilibrium" in which gene flow and drift balance each other and isolation by distance occurs as expected in a stepping-stone system. The second case involves a population too newly founded to exhibit equilibrium. Here all sub-population pairs will have approximately the same amount of differentiation despite distance (meaning the value of FST will be very similar for each pair) because of the founder effect and thus no correlation occurs between genetic and geographic distance. The third situation occurs when gene flow remains strong relative to drift so that there is a great degree of variation in the amount of differentiation between a given population pair, but there is no correlation with geographic distance. In the fourth and final case occurs when, at close distance gene flow still functions normally, allowing equilibrium, but due to environmental factors isolation occurs among some populations.
Darby Creek most closely resembles the third case. Standard deviation from the mean measure of genetic distance (FST = 0.037, Standard deviation = 0.030) is nearly equal to the mean itself and there is no correlation between genetic and geographic distance (r = 0.099). Levels of differentiation in L. siliquoidea are too variable to be due to recent founding effects, and given the small geographic scale it appears that gene flow and genetic drift are not at equilibrium, with gene flow influencing allele frequencies more than drift. Furthermore, all populations deviate from Hardy-Weinberg equilibrium at nearly all loci. Gene flow, which according to Nm seems barely high enough to prevent complete isolation, is actually easily sufficient at the geographic scale of the creek to overbalance genetic drift. The assignment test supported this hypothesis with a high proportion of incorrect assignment, suggestive of high mixing rates between populations.
Berg et al. (in press) suggested the ecological implication of this pattern. L. siliquoidea prefers lakes and small to medium-sized streams in mud, sand, or gravel (Cummings and Mayer 1992) and is usually found in fast deep (1-2m) water (Hart 1995). Berg et al. hypothesized that, since dispersal of unionid larvae is dependent on the movements of the host fish (Mulvey et al. 1998), measurements of genetic differentiation will be negatively correlated with host fish vagility. They further postulate that mussels from large river systems are likely to use larger, more mobile host fishes.
Q. quadrula in the Ohio River basin (Berg et al. 1998) is an example of this principle. Q. quadrula is known to parasitize flathead catfish (Pylodictus olivaris), which are known to traverse the entire river system (Ohio State University Museum of Biological Diversity (MBD) Mussel / Host Database; http://128.146.250.63/Musselhost/). This explains the lack of significant differentiation among populations. By comparison, L. siliquoidea's hosts include centrarchids and minnows (MBD) such as the smallmouth bass, Micropterus dolomei, which moves only short distances (Lyons & Kanehl 2002) making it unlikely for frequent exchanges between drainages or even streams (i.e. between Little and Big Darby creek). Small stream species are therefore highly likely to be evolutionarily distinct between drainages (King et al. 1999), a concept that has important conservation implications in terms of the habitat preference of target unionid species.
This study demonstrates significant levels of within-population variation as well as significant between-population differentiation. Given the small geographic scale, the level of gene flow exhibited by this species, and the results of Berg et al.'s (in press) earlier study using allozyme markers, it is hypothesized that L. siliquoidea will exhibit significantly more variation than demonstrated in species or habitats facing significant anthropogenic impacts. The present study observes that L. siliquoidea follows a non-equilibrium model similar to Wright's (1931) island model. The role of scale demonstrated here suggests the need for different conservation strategies to preserve genetic diversity in different unionid species.
Relocation and reintroduction of freshwater bivalves has not involved consideration of genetic information to date (Mulvey et al. 1998). Relocation generally may cause founder effects (including loss of heterozygotes) if too few individuals are moved with no consideration for genetics. In small populations, drift can cause rapid, random decreases in heterozygosity. Outbreeding depression, risk of spread of disease, and loss of local adaptations are consequences of relocation between isolated populations (Villella et al. 1998). Most important is the risk of loss of access to appropriate fish hosts. Ultimately, relocations or reintroductions need to be based on potential for gene exchange between geographic populations as well as data on host fishes and their availability in the area selected for colonization (Villella et al. 1998). In most cases, unionid relocations should be avoided between tributaries of the same drainage because these populations may have been reproductively isolated for thousands of generations. Therefore, unionid bivalves distributed among discontinuous habitats (e.g., Atlantic slope drainages) potentially should be considered evolutionarily distinct (King et al. 1999) and will therefore require separate management considerations in any given drainage.
ACKNOWLEDGEMENTS
This paper resulted from participation in a Research Experiences for Undergraduates (REU) program at Miami University, Oxford, Ohio entitled "Ecology in Human-Dominated Landscapes." Thanks to my Eckerd College committee, Drs. Peter A. Meylan (Chair), Steven Denison, and William A. Szelistowski, for their review of this manuscript, and to Elizabeth Ford for her assistance throughout this project. Financial support was provided by the National Science Foundation's REU Site Grant to Miami University (grant number DBI-0353915). This work was done as an undergraduate thesis in Biology at Eckerd College.
REFERENCES
Astanei, J, E. Gosling, J. Wilson, and E. Powell (2005) Genetic variability and phylogeography of the invasive zebra mussel, Dreissena polymorpha (Pallas). Molecular Ecology 14, 1655-1666.
Avise, J.C. (2004) Molecular markers, natural history, and evolution (2nd ed.). Sinauer Associates, Inc. Sunderland, MA.
Balloux, F. and N. Lugon-Moulin. (2002) The estimation of population differentiation with microsatellite markers. Molecular Ecology 11, 155-165.
Berg, D.J., A.D. Christian, and S.I. Guttman (In Press) Allozyme variation in freshwater mussel (Unionidae) species from small streams and the development of a hypothesis to predict genetic structure. Freshwater Biology.
Berg, D.J., E.G. Cantonwine, W.R. Hoeh, and S.I. Guttman. (1998) Genetic structure of Quadrula quadrula (Bivalvia: Unionidae): Little variation across large distances. Journal of Shellfish Research 17(5), 1365-1373.
Bogan A.E. (1993) Freshwater Bivalve Extinctions (Mollusca: Unionoida): a search for causes. American Zoologist 33(6), 599-609.
Bouzat, J.L., H.H. Cheng, H.A. Lewin, R.L. Westemeier, J.D. Brawn, and K.N. Paige (1998) Genetic evaluation of a demographic bottleneck in the greater prairie chicken. Conservation Biology 12, 836-843.
Burky, A.J. (1983) Physiological ecology of freshwater bivalves. In W.D. Russell-Hunter (Ed.), the Mollusca ecology (vol. 6; pp. 281-327), Academic Press Orlando, FL.
Cummings, K.S. and C.A. Mayer (1992) Field guide to freshwater mussels of the Midwest. Illinois Natural History Survey Manual 5.
Curole, J.P., D.W. Foltz, and K.M. Brown (2004) Extensive allozyme monomorphism in a threatened species of freshwater mussel, Margaritifera hembeli Conrad (Bivalvia: Margaritiferidae). Conservation Genetics 5, 271 278.
Davis, G.M. (1984) Genetic relationships among some North American Unionidae (Bivalvia): sibling species, convergence, and cladistic relationships. Malacologia 25(2), 629 648.
Duran, S., M. Pascual, A. Estoup, and X. Turon (2004) Strong population structure in the marine sponge Crambe crambe (Poecilosclerida) as revealed by microsatellite markers. Molecular Ecology 13, 511-522.
Eackles, M.S. and T.L. King (2002) Isolation and characterization of microsatellite loci in Lampsilis abrputa (Bivalvia: Unionidae) and cross-species amplification within the genus. Molecular Ecology Notes 2, 559-562.
EPA, Ohio (2002) Biological and water quality study of the Big Darby Creek watershed 2001/2002. Ohio EPA Technical Report EAS/2005-6-3.
Frankham, R. (1995) Conservation genetics. Annual Review of Genetics 29, 305-327.
Foltz, D.W. (1986) Null alleles as a possible cause of heterozygote deficiencies in the Oyster Crassostrea virginica and other bivalves. Evolution 40 (4), 869-870.
Fuller, S.L.H. (1974) Clams and mussels (Mollusca: Bivalvia). In Hart, C.W. and S.L.H. Fuller (eds.). Pollution ecology of freshwater invertebrates. pp. 215-274. Academic Press, New York.
Geist, J. and R. Kuehn (2005) Genetic diversity and differentiation of central European freshwater pearl mussel (Margaritifera margaritifera L.) populations: implications for conservation and management. Molecular Ecology 14, 425-439.
Hart, R.A (1995) Mussel (Bivalvia: Unionidae) habitat suitability criteria for the Otter Tail River, Minnesota: a thesis submitted to the graduate faculty of the North Dakota state University of Agriculture and Applied Science.
Hutchison, D.W. and A.R. Templeton (1999) Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution 53(6), 1898-1914.
Jass, J. and J. Glenn (2004) Sexual dimorphism in Lampsilis siliquoidea (Barnes, 1823) (Bivalvia: Unionidae). American Malacological Bulletin 18, 45-47.
Kat, P.W. (1983) Morphological divergence, genetics, and speciation among Lampsilis (Bivalvia: Unionidae). Journal of Molluscan Studies 49, 133-145.
Kelly, M.W. and J.M. Rhymer (2005) Population genetic structure of a rare unionid (Lampsilis cariosa) in a recently glaciated landscape. Conservation Genetics, 6:789-802.
Kimura, M. (1953) "Stepping-stone" model of population. Annual Report of the National Institute of Genetics Japan 3, 62-63.
Kimura, M. and G.H. Weiss (1964) The stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics, 49: 561-576.
King, T.L., M.S. Eackles, B. Gjetvaj, and W.R. Hoeh (1999) Intraspecific phylogeography of Lasmigona subviridis (Bivalvia: Unionidae): conservation implications of range discontinuity. Molecular Ecology 8, 65-78.
Liu H-P., J.B. Mitton, and S.J. Herrmann (1996) Genetic differentiation in and management recommendations for the freshwater mussel, Pyganodon grandis (Say, 1829). American Malacological Bulletin 13(1), 117-124.
Lyons, J. and P. Kanehl (2002) Seasonal movements of smallmouth bass in streams. In Philipp, D.P. and M.S. Ridgeway (eds.). Black Bass: Ecology, Conservation, and Management, Symposium 31. pp. 149-160. American Fisheries Society, Bethesda, Maryland.
Malécot, G. (1955) Remarks on the decrease of relationship with distance. Cold Spring Harbor Sympium on Quantitative Biology 20, 52-53.
Moritz, C. (1994) Defining evolutionary significant units' for conservation. Trends in Ecology and Evolution 9(10), 373-375.
Mulvey, M., C. Lydeard, D.L. Pyer, K.M. Hicks, J. Brim-Box, J.D. Williams, and R.S. Butler (1997) Conservation genetics of North American freshwater mussels Amblema and Megalonaias. Conservation Biology 11(4), 868-878.
Mulvey, M., H-P. Liu, and K.L. Kandl (1998) Application of molecular genetic markers to conservation of freshwater bivalves. Journal of Shellfish Research 17(5), 1395-1405.
O'Brien, S.J. and J.F. Evermann (1988) Interactive influence of infectious disease and genetic diversity in natural populations. Trends in Ecology and Evolution 3, 254-259.
O'Brien, S.J. and 10 others (1996) Conservation genetics in the Felidae. In Avise, J.C. and J.L. Hamrick (eds.). Conservation genetics: case histories from nature. pp. 50-74. Chapman and Hall, New York.
Peakall, R. and P.E. Smouse (2005) GenAlEx 6: Genetic Analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes In press (Available online at http://www.blackwell-synergy.com/loi/men).
Pianka, E.R. (ed.) (2000) Evolutionary Ecology. Adison Wesley Educational Publishers, Inc., San Francisco.
Quattro, J.M. and R.C. Vrijenhoek (1989) Fitness differences among remnant populations of the endangered Sonoran Topminnow. Science 245, 976-978.
Roe, K.J., P.D. Hartfield, and C. Lydeard (2001) Phylogeographic analysis of the threatened and endangered superconglutinate-producing mussels of the genus Lampsilis (Bivalvia: Unionidae). Molecular Ecology 10, 2225-2234.
Roe, K.J. and W.R. Hoeh (2003) Systematics of freshwater mussels (Bivalvia: Unionoida). In Lydeard, C. and D.R. Lindberg (eds.). Molecular systematics and phylogeography of mollusks. pp. 91-122. Smithsonian Books, Washington.
Saccheri, I., M. Kuussaari, M. Kankare, P. Vikman, W. Fortelius, and I. Hanski (1998) Inbreeding and extinction in a butterfly metapopulation. Nature 392, 491-494.
Sage, R.D. and J.O. Wolff (1986) Pleistocene glaciations, fluctuating ranges, and low genetic variability in a large mammal (Ovis dalli). Evolution 40(5), 1092-1095.
Serb, J.M. and C. Lydeard (2003) Complete mtDNA sequence of the North American freshwater mussel, Lampsilis ornata (Unionidae): an examination of the evolution and phylogenetic utility of mitochondrial genome organization in Bivalvia (Mollusca). Molecular Biology and Evolution 20(11), 1854-1866.
Slatkin, M. (1987) Gene flow and the geographic structure of natural populations. Science 236(6), 787-791.
Slatkin, M. and N.H. Barton (1989) A comparison of three indirect methods for estimating average levels of gene flow. Evolution 43(7), 1349-1368.
Spielman, D., B.W. Brook, and R. Frankham (2004) Most species are not driven to extinction before genetic factors impact them. Proceedings of the National Academy of Sciences of the United States of America 101, 15261-15264.
Stiven, A.E. and J. Alderman (1992) Genetic similarities among certain freshwater mussel populations of the Lampsilis genus in North Carolina. Malacologia 34, 355-369.
Strayer, D.L., J.A. Downing, W.R. Haag, T.L. King, J.B. Layzer, T.J. Newton, and S.J. Nichols (2004) Changing perspectives on pearly mussels, North America's most imperiled animals. Bioscience 54(5), 429-439.
Turgeon D.D., J.F. Quinn, and A.E. Bogan Jr. (1988) Common and scientific names of aquatic invertebrates from the United States and Canada: mollusks (2nd ed.) American Fisheries Society Special Publication 26, American Fisheries Society Bethesda, MD.
Turner, T.F., J.C. Trexler, J.L. Harris, and J.L. Haynes (2000) Nested cladistic analysis indicates population fragmentation shapes genetic diversity in a freshwater mussel. Genetics 154, 777-785.
Vaughn, C.C. and C.C. Hakenkamp (2001). The functional role of burrowing bivalves in freshwater ecosystems. Freshwater Biology 46, 1431-1446.
Villella, R.F., T.L. King, and C.E. Starliper (1998) Ecological and evolutionary concerns in freshwater bivalve relocation programs. Journal of Shellfish Research 17(5), 1407-1413.
Waser, P.M., D. Paetkau, and C. Strobeck (2001) Estimating interpopulation dispersal rates. In Gittleman, J.L., S.M Funk, D.W. Macdonald, and R.K. Wayne (eds.) Carnivore Conservation. pp. 484-497, Cambridge University Press, Cambridge.
Watters, G.T. (1996) Small dams as barriers to freshwater mussels (Bivalvia, Unionoida) and their hosts. Biological Conservation 75, 79-85.
Wayne, R.K., N. Lehman, D. Girman, P.J.P. Gogan, D.A. Gilbert, K. Hansen, R.O. Peterson, U.S. Seal, A. Eisenhawer, L.D. Mech, and R.J. Krumenaker (1991) Conservation genetics of the endangered Isle Royale gray wolf. Conservation Biology 5(1), 41-51.
Williams, J.D., M.L. Warren Jr., K.S. Cummings, J.L. Harris, and R.J. Neves (1993) Conservation status of freshwater mussels of the United States and Canada. Fisheries 18(9), 6-22.
Wright, S. (1931) Evolution in Mendelian populations. Genetics 16, 97-159.
Wright, S.(1943) Isolation by distance. Genetics 28, 114-138.
Wright, S. (1951) The genetical structure of populations. Annals of Eugenics 15, 323-354.
Young, D.L., M.W. Allard, J.A. Moreno, M.M. Miyamoto, C.R. Ruiz, and R.A. Perez-Rivera (1998) DNA fingerprint variation and reproductive fitness in the plain pigeon. Conservation Biology 12(1), 225-227.