Author: Tony Tu
Institution: Duke University
Date: November 2005
Abstract
Pharmacogenomics is the study of genetic differences responsible for the variability in drug response among individual patients. Emerging from this new science is the development of diagnostic tools crucial for decreasing drug side effects and optimizing treatment strategies. However, both application and progress have been troublesome in guiding treatment of genetically-linked diseases such as cancer and diabetes. The complexity of multiple gene interactions requires a collaborative effort to understand the genetic bases of variable drug response. Societal quandaries also burden research. The novel genomic information to be used in tackling disease may be viewed as discriminatory factors involving winners and losers. Concerns of privacy and cost as well as changes to today's medical education and practice will unsettle the American healthcare system. This article reviews the feasibility and potential of applying the latest advancements in genomics to medicine and drug development. Examples of prevalent diseases and polymorphisms are discussed as avenues in which pharmacogenomics have relevance. In addition, the fears of race-based medicine will be clarified with regards to the future of this new science, with supporting lines of evidence on how race is one factor among many that reveals the true potential of pharmacogenomics in guiding a doctor's decision.
Introduction
Pharmacogenomics is the study of how an individual's genome influences his or her response to a specific drug. Conceived five years after the Human Genome Project had been erected, the notion of pharmacogenomics has solidified into an emerging science that will greatly impact the future of medicine. Pharmacogenomics will systematically change the design of today's drug pipeline by streamlining drug development. For example, decreasing the size of clinical trials and screening for feasible drug targets among patients will boost the number of drugs that reach the market and replenish today's lackluster arsenal of medications. Once implemented, pharmacogenomics is hoped to boost drug efficacy by guiding physicians to the best drugs and optimal prescriptions for different patients (Vascar et al., 2002).
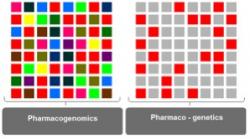
Figure 1. A model for understanding pharmacogenomic and pharmacogenetic approaches towards studying variable drug response. Individual squares represent genes that affect drug response. The entire 8X8 ensemble represents 64 genes involved in determining drug response. Each color represents one underlying mechanism. Red may represent drug metabolism; blue, drug transport; green, drug excretion; etc. Squares of the same color represent all the genes involved in one particular mechanism. Pharmacogenomics considers all 64 squares (i.e., all mechanisms and the respective genes). Pharmaco-genetics approaches drug response one mechanism at a time or one gene at a time. In order to understand drug response, both the big picture (all 64 squares) at the level of the genome as well as the fine details at the level of the allele must be considered. Thus, the two disciplines are tightly intertwined, and nomenclature is often interchangeable, with pharmaco-genomics often representing both sciences. In the clinical setting, pharmacogenomics can (1) identify patients at high risk for adverse drug responses and (2) optimize therapy for each patient by tailoring medicine. In addition, pharmacogenomics can speed up drug development by selecting feasible drug targets, reducing clinical trial sizes, and increasing safety of the trials.
The basis of pharmacogenomics is founded on pharmacogenetics, the study of individual genetic differences (polymorphisms) in drug absorption, metabolism, distribution, and excretion. The two terms are often used interchangeably as both are closely intertwined in studying drug response (Figure 1). Beginning in the early 1950's, pharmacogenetics has focused on the link between polymorphic genes and adverse drug responses. Pharmacogenetics focuses on variable drug responses arising from polymorphisms by individuals. The clearest examples involve polymorphisms in single genes that alter drug transport, metabolism, or clearance. Progress in pharmacogenetics is aimed at the clinical setting to find the best drug and optimal dosage for at-risk patients (Lindpainter, 2002).
From the early 1950s until a few years ago, progress was quite limited. The available technology allowed for only tiny snapshots of an individual's genome. During this period, many adverse drug responses were found to be genetically linked. Most prominent among these complications has involved the anti-cancer drugs thioguanine and mercaptopurine. These thiopurines resemble the guanine nucleotide and interfere with DNA replication. The enzyme thiopurine methyl-transferase (TPMT) metabolizes these agents. However, people who are homozygous for the null TPMT allele will acquire acute myelosuppression if they take the thiopurine drugs, mercaptopurine and azathiopurine. Less than half of a percent of the general population have this genetic disposition and should avoid thiopurine drugs at all costs (Vascar et al., 2002).
Very few diseases, however, involve single genes. Pharmacogenomics extends this gene-by-gene view to a whole-genome level, considering not only multigenic scenarios but also how multiple drugs can alter genome expression. Rather than focusing on genetic variability among patients, emphasis is placed on better prediction of drug toxicities, screening out drugs with complications, and identification of the best drug candidates (Lindpaintner et al., 2002). The approaches of these two areas in improving medicine are complimentary. Pharmacogenetics begins by examining the differences among patients, while pharmacogenomics begins by examining the differences between drugs.
Figure 2. Genetic polymorphisms can alter individual drug responses by interfering with drug metabolism. By identifying patients with polymorphisms that negatively interact with specific drugs, physicians can avoid harmful side-effects and optimize therapy.
Among the approximately 30,000 genes in the human genome (Brentani et al., 2003), variations in the form of single nucleotide polymorphisms (SNPs) largely account for the variabilities among patients in their responses to drugs. SNPs alter genes encoding proteins essential for drug metabolism (Lee et al., 2005). Many enzymes that have crucial roles in drug metabolism have high frequencies of polymorphisms among individuals in the human race (Figure 2).
In the past, drug industries have avoided polymorphic enzymes due to the high variability in drug response (Walker, 2004). However, the latest genomics research has brought to drug development the potential to match patients with the most suitable drug based on polymorphisms.
Cancer is the paradigm disease in which high variability in drug responses complicate treatment. Pharmacogenomics can advance cancer drug research and provide extensive preliminary information about patients in clinical trials (Lee et al., 2005). Because the drugs used in chemotherapy tend to have very narrow therapeutic indices (i.e., very little room for error in dosage), individualized chemotherapy regimens can reduce the fatal side-effects of imprecise dosage. Drug side-effects, commonly referred to as "adverse drug reactions" (ADRs), are the fifth leading cause of death in hospitalized patients in the U.S. as of, 2000. Examples of ADRs include increased pain, higher frequencies of emergency room visits, and additional hospitalizations and prescriptions, all of which raise healthcare costs (Bukaveckas, 2004).
Pyrimidine antagonists (PAs), a class of chemotherapy drugs including gemcitabine and 5-fluorouracil, was recently found to have high variability in patient response (Maring et al., 2005). Used to treat a range of cancers including pancreatic, breast and lung cancers, these drugs act as antagonists against DNA precursor molecules and interfere with DNA replication similar to the thiopurine drugs mentioned earlier. In order to achieve cytotoxic effects, these drugs must first be metabolized into active compounds. Polymorphisms in the genes responsible for these metabolizing enzymes largely differentiate treatment outcomes. Some patients benefit more, while others benefit less from identical chemotherapy regimens (Maring et al., 2005). Allele frequencies for PA polymorphisms range from less than 1% to more than 95%, particularly among different ethnicities. For example, one polymorphism for thymidylate synthase whose inhibition is a major target of fluorouracil occurs at an allele frequency of 2641% in Caucasians and 76% in Singaporean Chinese (Maring et al., 2005).
However, experts have concluded that the PA polymorphisms discovered thus far cannot serve as reliable markers for predicting treatment efficacy in different patients because larger studies yielding greater statistical power are needed to verify the specificity of these potential markers (Maring et al., 2005). Nevertheless, the long list of potential polymorphisms may be useful for devising large scale genotyping, or screening panels of genes for polymorphisms of not only single but also multiple genes.
Advances in the Field
Pharmacogenomics can help overcome two prominent factors responsible for complications of today's cancer chemotherapy. First, patients usually have variable responses to chemotherapy, muddling decisions as to which drugs or cocktail of drugs will be optimal. Second, the arsenal of drugs used in chemotherapy today usually has very narrow therapeutic indices (Lee et al., 2005).
Application of pharmacogenomics can address therapeutic indices by predicting patient response and speeding up the rate of drug development and clinical trials (Lee et al., 2005). Recognizing the polymorphisms associated with disease makes individualized treatments possible, which can lower risks of imprecise dosage.
Among the many diseases associated with variability in drug response, cancer has benefited the most from pharmacogenomics research. While environment, age, diet, and other external factors are important, hereditary aspects of cancer have also been found to contribute to the variability in drug response of patients undergoing chemotherapy (Lee et al., 2005). The inherited genetic polymorphisms are associated with malfunctioning enzymes, including drug metabolizers, and take the form of DNA sequence mutations (deletions, repeats, insertions) and SNPs.
A prominent example of how such a polymorphism can result in a higher patient susceptibility to ADRs is found in the treatment of leukemia. As described earlier, patients genetically endowed with weakened versions of TPMT cannot receive the regular dosage of thiopurine agents that interfere with DNA replication in rapidly dividing cells. Leukemia patients with a TPMT polymorphism require a lowered dosage to prevent toxic side-effects because not enough TPMT is present to metabolize the drug (Lee et al., 2005; Lennard et al., 1993).
Methods have been established to diagnose TPMT deficiency to address this identifiable risk in leukemia patients undergoing chemotherapy, and physicians are advised to use an alternative therapy or to reduce dosage levels thiopurine drugs (Lee et al., 2005). This diagnostic step is particularly important for children with acute lymphatic leukemia, as thiopurine dosage optimization has been directly correlated with higher survival rates (Lee et al., 2005, Lennard et al., 1993). As long-term chemotherapy can have many detrimental effects, a major project lead by researchers in Australia aims to apply pharmacogenomic tools to the risk assessment of children undergoing chemotherapy in order to establish a highly specific diagnostic ability. While screening for TPMT polymorphisms is quite logical, the genotype alone is inadequate for optimizing treatment because thiopurine metabolism is not simply black and white. Patients with completely normal alleles may require reduced dosage, and vice versa (Zineh et al., 2004).
For lung cancer, testing levels of mRNA expression of relevant genes now provide information about patients that helps doctors decide whether to use gemcitabine or pemetrexe, two different chemotherapy cocktails, alone or in combination (Rosell et al., 2004).
Similar approaches have been adopted in other diseases such as asthma, in which several polymorphisms have been associated with variable responses from patients using the three major drug types: glucocorticosteroids, leukotriene modifiers, and beta-agonists (Sayers et al., 2005). Researchers who approach asthma patients located in places where such polymorphisms are prevalent will have the capacity to test, identify, and match specific patients to specific treatment plans. Pharmacogenomic tests are thus a major supplement for established diagnostic measures that are based on mere phenotypic features such as weight or levels of specific biomarkers in the blood (Rosell et al., 2004).
Future Research Areas
The technology for genomics research has become increasingly automated, less expensive, and more rapid, allowing for greater capacity to implement genetic tests with high predictive values for drug development and clinical care. (Lee et al., 2005). As today's genetic analyses become increasingly comprehensive, an ever expanding database of SNPs is at scientists' disposal for the matching of alleles to diseases the first step in understanding variabilities in drug response.
What is necessary now for genetic analysis of SNPs is the rapidly detection and discrimination of SNPs in patient samples. The polymerase chain reaction (PCR) is a highly effective tool for amplifying regions of the genome containing the desired SNPs, and many innovations have been implemented to increase the amplification capacity of PCR (Kwok et al., 2003; Kennedy et al., 2003).
The latest innovation in PCR technology allows the simultaneous analysis of over 1,000 SNPs using haploid genetic material, or DNA from sperm and eggs (Wang et al., 2005). In addition to increasing the throughput of large-scale genetic analyses, this new system requires a considerably less amount of DNA than previous systems, making tests more practical on highly preserved samples which have low, degraded amounts of DNA, such as tissues from cancer biopsies. In short, the improvements of this new technology, including a simpler protocol, higher sensitivity, no specialized equipment, and fewer reagents, greatly facilitate large-scale SNP analyses on samples that were previously impossible.
The technology for measuring levels of gene expression have also advanced rapidly, allowing for simultaneous measurements in the form of high-density oligonucleotide arrays (Ronald et al., 2005). This technology for exploring entire genomes can study a much larger quantity of genes and also requires simpler procedures than previous methods, making it ideal for identifying candidate genes of complex diseases.
Such lines of technology can provide the genetic information for making predictions that associate specific polymorphisms in a patient with a specific drug metabolizing enzyme or drug target (Lee et al., 2005). Should that polymorphism be linked with a deficit, the patient may then be steered to a better route of treatment.
Concerns of Race-Based Medicine
Some polymorphisms that raise variability in drug metabolism occur at higher frequencies in the genomes of particular races. The P-glycoprotein (PGP) is a transporter that distributes many types of drugs, including those used in chemotherapy. Globally, there is an immense genetic variation in the gene encoding PGP. What is interesting in the case of PGP is that different races have been characterized as having higher frequencies of certain PGP polymorphisms. Studies have shown that African people have about an 80% chance of having one specific version while Asian and European peoples have half that chance (Kim et al., 2001).
The advent of BiDil marks the first drug in America to be marketed exclusively for African Americans. Data from the trial that studied the effects of BiDil on congestive heart failure among African American patients showed a 43% improvement in survival rate (Taylor et al., 2004).
NitroMed, the maker of BiDil, had originally tested the components of BiDil in the general population several years ago. The company's recent finding showed significant efficacy within a subpopulation of African Americans (Taylor et al., 2004). The company announced early February of this year that the U.S. Food and Drug Administration (FDA) had accepted their drug for resubmission.
The controversy lies in how the notion of race is used. For physicians, what matters is the 43% increase in efficacy. However, they will have to make decisions based on the patients' race. Dr. Robert Cook-Deegan, the director of the Institute for Genome Sciences and Policy's (IGSP) Center for Genome Ethics Law and Policy, posed the following question to his law and ethics class at Duke. "Say you're the physician. A new patient walks in. You don't know anything about them, but they've got congestive heart failure. What are you going to do? Are you going to ask them what race they are or are you going to make your own judgment about whether or not they are African Americans?" This is the dialogue that doctors will be having with their patients.
Indeed, the development of BiDil and other future drugs tailored to subgroups is a good thing in that it works well for people. Yet the next logical step for researchers would be to determine if there is a genetic factor. Dr. Cook-Deegan noted the possibility that BiDil's special efficacy in African Americans may not be a genetic factor at all.
"We have an association between self-designated race and the use of this drug, but what we don't have is the causal relationship that proves why this is," said Dr. Cook-Deegan. "The fact that there's some association with race does not mean that it's biological. For the drugs that seem to have higher efficacy or safety profiles in specific races, there are many existing factors such as the environment, dietary habits, and physiology."
It would be necessary to find what determines patient response to BiDil and to individualize treatment, noted Dr. David Goldstein, Wolfson Professor of Genetics at University College London, now the director of the Center for Pharmacogenetics and Population Genomics at the IGSP.
"It can only be an interim solution to use the drug as an ethnically indicated medicine. In the long term, that's a mistake because that will serve to reinforce the notion that racial and ethnic groups are distinct groups, and they most certainly are not," said Dr. Goldstein.
The issue raised by BiDil's racially profiling nature is that race is not a marker that can be used as a definitive basis for making prescriptions. The study that showed the 43 percent relative decrease in heart failure in the African American group had patients who were "self-identified" as blacks (Taylor et al., 2004). According to a review by Dr. Gregg Bloche, such an inclusion separated this study from the "deeper" science of population genetics because people who may not have had any black ancestors but still considered themselves black were included in the study (Bloche, 2004). Furthermore, Bloche noted that even among Americans of African descent, the genetic heterogeneity of the population on the African continent is far more diverse than in other races. Should the same BiDil study be carried on random sample in Africa, results may be considerably varied.
"In the end there's not going to be one pill for a particular race. The point is that the variations, more often than not, will be in all groups. It's just that their distribution will be different," said Dr. Marchuk, a molecular geneticist and co-director of Duke's Program in Genetics. "By and large, we're going to find that genetic variation does differ among groups. But the pill' isn't for one race. It's for whoever has that variant."
Race for Pharmacogenomics
Although at best a yardstick, race has been an important factor in risk assessment for side-effects and variability in drug response. Moreover, innovations from a pharmacogenomic perspective have been greatly facilitated based on racial associations. The following diseases are examples of cases where consideration of race has been played a strong role in directing advances in healthcare and the applicability of pharmacogenomics.
Breast cancer. BRCA1 and BRCA2, two of the genes associated with the onset of breast cancer, have several polymorphisms identified (McClain et al., 2005). In particular, three polymorphisms in these two genes are approximately ten times more prevalent in Ashkenazi Jewish women than in American women (McClain et al., 2005). The risks for developing breast cancer are drastically increased given the presence of these mutations. A mathematical equation has recently been devised to assess the magnitude of the risk for developing breast cancer given the presence of BRCA1 and BRCA2 mutations, integrating several disease parameters that had never before been studied together (McClain et al., 2005). This risk assessment has refined current diagnostic capacities to implementing interventional approaches as early as possible.
Type-2 diabetes mellitus. The primary defect responsible for this disease is varied among populations, especially in Japanese and Caucasians. As the development of type-2 diabetes can arise from two different paths (i.e., impaired insulin secretion or impaired insulin sensitivity), recent studies have shown that diabetes in Japanese people arise primarily through problems in secretion whereas Caucasian diabetics primarily have problems with sensitivity (Martin et al., 1992). This difference was explored genetically by Sasazuki et al., who identified several genetic loci linked to development of type-2 diabetes. This was a major step towards establishing personalized medicine for Japanese people (2004). This genomic study follows and confirms two previous projects associating occurrence of type-2 diabetes in Japanese people to specific regions in the genome susceptible for the unique mode of disease development.
Phenylketonuria (PKU). PKU is an autosomal recessive metabolic condition that has been used as a classic model for teaching hereditary diseases in biology. The occurrence of PKU varies among different races (1/10,000 in Caucasians, 1/120,000 in Japanese, and 1/41,000 in Koreans) (Bickel et al. 1981, Aoki et al. 1988, and Lee et al., 2004). The gene for phenylalanine hydroxylase (PAH), whose defective alleles are responsible for PKU, was recently examined among Chinese, Japanese, and Korean people (Lee et al., 2004). In comparing the mutations and polymorphisms in the PAH gene among these three races, several features were found to be different in the Korean genetic profile for PAH. Because the severity of PKU is diverse and treatment options can range from dietary regimens to prescribed medications, identifying the polymorphisms underlying the disease may provide tailored information for diagnosis, treatment plans, and genetic counseling for Chinese, Japanese, and Koreans with PKU.
Colorectal cancer. Fluoropyrimidines, a popular chemotherapy agent used to treat colorectal cancer, kills tumors by interfering with DNA synthesis (Wiemels et al., 2001). The drug must first be activated by binding to methylenetetrahydrofolate (MTHF), a cellular molecule present at varying concentrations among individuals. Effective killing of tumors requires high concentrations of MTHF. Patients deficient in MTHF usually receive supplements prior to chemotherapy. However, what ultimately determines the treatment efficacy is the activity of the enzyme methylenetetrahydrofolate reductase (MTHFR), which controls the rate of synthesis of MTHF. The MTHFR gene is known to have several polymorphisms among populations. In particular, one single nucleotide polymorphism is 10-16% among Caucasians but only a few percent among African-Americans (Wiemels et al., 2001). Different MTHFR polymorphisms confer different levels of sensitivity to the chemotherapy (Etienne et al., 2004). Greater drug efficacy was found in tumor cells with particular MTHFR mutations, confirming the effectiveness of treatment options based on genetic information (Etienne et al., 2004).
Consideration of a patient's race does carry weight in assessing risks for drug side-effects. But what scientists have found on polymorphisms associated with race and drug response is only a prelude to the future of a genome-based medicine. With regard to the health disparities associated with race, Dr. Goldstein noted that more than 29 drugs have been claimed by various literatures to have different levels of efficacy among different racial groups (Goldstein et al., 2004). For now, the most efficacious approach is not to reject the use of race as a diagnostic tool, nor to rest in complacency at the improved results as in the case of BiDil. Genetic differences among racial groups do exist and are usually related to environmental factors (Goldstein et al., 2004). What is important to realize is that "race" is an abstract descriptor that accounts for environmental differences and other factors outside of the genetics realm. From a pharmacogenomics perspective, improving drug efficacy should not strictly entail genomic technologies, but also include established practices of medicine.
Pharmacogenomics Today
While benefits from pharmacogenomics have yet to fully reach the patient, small-scale genetic tests have surfaced. Last year, a comprehensive analysis of pharmacogenomics-related data in drug package inserts found that pharmacogenomics information was present in very few inserts (Zineh et al., 2004). Not even 18% of the drugs were in the top, 200 drugs of, 2002. The authors concluded that many of these inserts did not contain enough information for adequate guidance on dosage or therapy. Moreover, the authors found that as many as half of the genes currently listed in drug package inserts play roles in drug distribution, transport, excretion, and pathways other than direct drug metabolism. Screening for individual polymorphisms and interpretation of the resulting data is much more complicated for most diseases today. Furthermore, most diseases are marked not by single genes but by diverse arrays of genes, calling for whole genome approaches (Zineh et al., 2004).
Yet the information gained by whole genome approaches will reveal exponentially more information than simply risks for adverse drug responses. Stored in enormous genomic databases, the sheer magnitude of the number of genes used for screening genomes poses many problems for information privacy. With each bit of genetic fact found about a patient, the likelihood of unintended facts being revealed increases due to pleiotropy, in which a polymorphism of one gene can affect many phenotypic characters (Vascar et al., 2002). Family and close relatives, who have a higher risk of carrying the same polymorphisms as the patient, will also be unintentionally influenced by the new information gained from screenings.
Whether the new information conveys an increased risk for diabetes or Alzheimer's, the quality of life for the patient and others close to him or her may be subjected to non-physical harm, with new anxieties and implied limitations. Businesses and insurance agencies will likely favor clients endowed with good genotypes in order to minimize genetically predicted overhead costs and liabilities (Vascar et al., 2002). These issues are only a sampling of the concerns beginning to surface in the wake of pharmacogenomics, all of which need to be addressed for the sake of the patient who could have been given something better.
References
Bloche, M.G. (2004). Race-based therapeutics. New England Journal of Medicine. 351: 2035-2037.
Etienne MC, Formento JL, Chazal M, Francoual M, Magne N, Formento P, Bourgeon A, Seitz JF, Delpero JR, Letoublon C, Pezet D, Milano G. (2004). Methylenetetrahydrofolate reductase gene polymorphisms and response to fluorouracil-based treatment in advanced colorectal cancer patients. Pharmacogenetics. 14: 785-92.
Goldstein, D.B. and Tate, S.K. (2004). Will tomorrow's medicines work for everyone? Nature: Genetics. 36: S34-S42.
Kim, R.B., Leake, B.F. and Choo, E.F. Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clin Pharmacol Ther. 70: 189-199.
Lee, D.H., Koo, S.K., Lee, K., Yeon, Y., Oh, H., Kim, S., Lee, S., Kim, S., Lee, J., Jo, I. and Jung, S. (2004). The molecular basis of phenylketonuria in Koreans. J Hum Genet. 49: 617-621.
Lee, W., Lockhard, C., Kim, R.B. and Rothenberg, M.L. (2005). Cancer pharmacogenomics: powerful tools in cancer chemotherapy and drug development. The Oncologist. 10: 104-111.
Lennard, L. Gibson, B.E. and Nicole, T. (1993). Congenital thiopurine methyltransferase deficiency and 6-mercaptoppurine toxicity during treatment for acute lymphoblastic leukaemia. Arch Dis Child. 69: 577-579.
Lindpaintner, K. (2002). The impact of pharmacogenetics and pharmacogenomics on drug discovery. Nature Reviews: 463.
Maring, J.G., Groen, H., Wachters, F.M., Uges, D., and de Vries E.G. (2005). Genetic factors influencing Pyrimidine antagonist chemotherapy. The Pharmacogenomics Journal: 226-243.
McClain MR, Nathanson KL, Palomaki GE, Haddow JE. (2005). An evaluation of BRCA1 and BRCA2 founder mutations penetrance estimates for breast cancer among Ashkenazi Jewish women. Genet Med. 7: 34-39.
McClain MR, Palomaki GE, Nathanson KL, Haddow JE. (2005). Adjusting the estimated proportion of breast cancer cases associated with BRCA1 and BRCA2 mutations: public health implications. Genet Med. 7: 28-33.
Nawata H, Shirasawa S, Nakashima N, Araki E, Hashiguchi J, Miyake S, Yamauchi T, Hamaguchi K, Yoshimatsu H, Takeda H, Fukushima H, Sasahara T, Yamaguchi K, Sonoda N, Sonoda T, Matsumoto M, Tanaka Y, Sugimoto H, Tsubouchi H, Inoguchi T, Yanase T, Wake N, Narazaki K, Eto T, Umeda F, Nakazaki M, Ono J, Asano T, Ito Y, Akazawa S, Hazegawa I, Rosell, R. Taron, M. Sanchez, J.M., Moran, T., Reguart, N., Besse, B., Isla D., Massuti, B., Alberola, V. and Sanchez, J.J. (2004). The promise of pharmacgoenomics: gemcitabine and pemetrexe. Oncology. 18: 70-76.
Sayers, I. and Hall, I.P. (2005). Pharmacogenetic approaches in the treatment of asthma. Curr Allergy Asthma Rep. 5: 101-108.
Taylor, A.L., Ziesche, S., Yancy, C., Carson, P., D'Agostino, R., Ferdinand, K., Taylor, M., Adams, K., Sabolinski, M., Worcel, M. and Cohn, J.N. (2004). Combination of Isosorbide Dinitrate and Hydralazine in Blacks with Heart Failure. The New England Journal of Medicine. 351: 2049-2057.
Takasu N, Shinohara M, Nishikawa T, Nagafuchi S, Okeda T, Eguchi K, Iwase M, Ishikawa M, Aoki M, Keicho N, Kato N, Yasuda K, Yamamoto K, Sasazuki T. (2004). Genome-wide linkage analysis of type 2 diabetes mellitus reconfirms the susceptibility locus on 11p13-p12 in Japanese. J Hum Genet. 49: 629-634.
Vascar, L.T., Rosen, G.D., and Raffin, T.A. (2002). Pharmacogenomics and the challenge to privacy. The Pharmacogenomics Journal: 144-147.
Walker, D.K. (2004). The use of pharmacokinetic and pharmacodynamic data in the assessment of drug safety in early drug development. Br J Clin Pharmacol. 58: 601-608.
Wang, H.Y., Luo, M., Tereshchenko, I.V., Frikker, D.M., Cui, X., Li, J.Y., Hu, G., Chu, Y., Azaro, M.A., Lin, Y., Shen, L., Yang, Q., Kambouris, M.E., Gao, R., Shih, W. and Li, H. (2005). A genotyping system capable of simultaneously analyzing >1000 single nucleotide polymorphisms in a haploid genome. Genome Res. 15: 276-283.
Zineh, I., Gerhard, t., Aquilante, C.L., Beitelshees, A.L., Beasley, B.N., and Hartzema, A.G. (2004). Availability of pharmacogenomics-based prescribing information in drug package inserts for currently approved drugs. The Pharmacogenomics Journal: 354-358.