Author: Anna E. Ritchie
Institution: Hawaii Pacific University
Date: April 2005
Abstract
In the last few years, the severity of coral bleaching events and disease observances has increased worldwide. Very little is known about disease in the Hawaiian Islands, although it is a commonly held belief that conditions are generally unfavorable for disease development/establishment due to Hawaii's open ocean circulation and low human population. This study focuses on characterizing bacterial communities found on healthy and lesioned Montipora capitata, Porites compressa, and Porites lobata collected from various sites around Oahu, Hawaii including the North Shore, Hanauma Bay, Kaneohe Bay and Kahe Point. Isolation of bacterial communities from the surrounding water, coral mucus layer, and the water used for transporting the coral were identified using partial 16S rDNA sequencing. As reported in other corals using similar methods, it was found that mucus from healthy and diseased M. capitata evidenced some differential composition of bacterial communities, while bacterial communities isolated from the surrounding waters were found to be independent of the bacterial communities in the mucus, suggesting microbe-coral specific associations. The bacterial communities associated with healthy tissues were also investigated in order to establish a baseline for Hawaiian corals. These results will contribute towards developing a standard for monitoring coral disease by comparing shifts in bacterial communities.
Introduction
Coral reefs play critical roles in marine ecology by providing vertebrates and invertebrates a place to live, reproduce, find refuge, and find food. Although many coral reefs have been in existence for thousands of years, they remain a very fragile system. Certain corals like Porites species and Acroporidae species are principal reef builders and are dominant species within the coral community, providing a three-dimensional habitat for many animals (Hoover, 1998). Recent observations indicate increasing sea temperatures and sea levels are having major impacts on the health of coral reefs, in some cases even shifting coral reef-dominated systems into algae-dominated systems (Harvell et al. 1999).
Corals have been shown to have associations with bacteria in the mucus layer and leading edge of coral tissues; whether this is simply uptake from the water column or is a source of biological activity remains uncertain (Ritchie and Smith 1997; Rohwer et al. 2001). The bacterial communities associated with the mucus layer are thought to vary compared to the surrounding waters. Frias-Lopez and collaborators (2002) have found that bacterial communities associated with healthy corals are distinctly different from the bacterial communities in their immediate surrounding environment. A study on the Caribbean coral Montastraea franksii, has been shown to possess a microbial community that is distinct from the surrounding environment, as well as possessing a specific association with an Alpha proteobacterium not found in the surrounding environment (Rowher et al. 2002). That said, while mounting evidence is suggestive, there are relatively few studies that confirm that coral species have specific bacterial associations. One study by Ritchie and Smith (1997) studied carbon source utilization patterns to demonstrate that coral-specific bacterial associations depend upon the structure of the mucus layer. Together, these studies highlight the fact that identifying the normal microbial community of coral species can aid scientists to understand the role that bacteria serve in maintaining healthy coral surfaces and highlight how changes in surface microbial communities may cause or reflect the onset of disease. As corals become stressed, mucus production increases, and the microbial community concomitantly changes in overall numbers and composition (Ritchie and Smith 1997).
Even though coral disease has been a topic of study for approximately 30 years, very little is known. Extant research has primarily focused on etiologic agents of disease and discovering whether there is truly a pathogenic bacterium involved in various coral diseases or whether environmental stressors contribute to opportunistic bacterial infections. The severity of coral disease depends on the rate at which the etiologic agent spreads, and environmental factors such as elevated temperature and increased nutrients. For example, in 1980, a fast-moving outbreak of white band disease in Acropora, species devastated Caribbean and Florida reefs (Harvell et al., 1999). While white band only affects Elkhorn corals, it has significantly reduced their numbers in these waters, which has undoubtedly disturbed the ecological balance of the overall reef.
Many factors such as increased sedimentation, human sewage, and global warming have been linked to the emergence of coral disease, but how these interplay with other factors that are thought to affect coral disease is still very uncertain (Sutherland et al., 2004). Efforts to characterize the bacterial communities in healthy tissues are undeveloped, as are efforts to identify disease-causing bacteria. To date, disease causation has only been confirmed in black band, white band, some examples of bacterial bleaching, coral plague, serratiosis (white pox), and aspergillosis (Rosenberg, E. and Ben-Haim, Y., 2002; Sutherland et al., 2004). Much of the research that has been accomplished has focused on Caribbean and Mediterranean regions with little focus on disease in the Hawaiian Islands. Part of the reason for this has been a preconceived notion that due to its open ocean circulation and low human population, the Hawaiian environment is less susceptible to disease caused by reduced water circulation or anthropogenic effects. Unfortunately, coral lesions (defined as growth anomalies, discoloration, and bleaching) are increasingly observed in Hawaiian waters (personnel communication, Greta Aeby, Hawaii DAR) and consequently, a need to address this issue is now recognized.
This study characterizes bacterial communities associated with surface microbes present in Porites compressa, Porites lobata, and Montipora capitata using molecular and microbiology culture techniques. These are the primary corals found in the main Hawaiian Islands. The physical structures of finger-like, knobby projections offered by P. compressa, may provide a different microhabitat than the more flat lobed coral P. lobata,, and may therefore affect the types of bacteria that colonize these two corals. Different physical structures may affect factors like different rates and spread of the mucus layer, which has been said to affect bacterial community composition (Pantos et al., 2003). In addition, preliminary disease assessment of bacterial communities present in healthy and diseased M. capitata tissues will be attempted, to identify non-mucus associated microbes. This was possible due to a larger number of M. capitata samples collected and sample types processed during this initial phase of the study.
Methods & Materials
Sample Collection
Figure 1. ArcView map of Oahu, Hawaii illustrating sample sites at Kaneohe Bay, Kahe point, Hanauma bay, and North shore.. The CD markings indicate individual sample sites. Lea Hollingsworth at HIMB created this map.
Coral samples were collected by AAUS certified divers with expertise in visual examination of coral health from various sites around Oahu (Figure 1). The sampling strategy was to collect nubbins of all species that displayed abnormalities and for each lesioned sample collected, a paired healthy coral nubbin was collected as a control. It was determined that depending on the number of samples collected for each species during the surveys, the disease dataset would include the species with the highest collection number (n) for statistical reasons. Paired coral nubbins were transported and processed on the same day as collection at the Hawaii Institute of Marine Science. The three sample types in this study included (in order of collection), seawater collected above the coral, coral mucus, and a water sample from bags used to transport the coral nubbins to the lab (containing water and coral mucus sloughed during transport). During sampling, the species of coral was recorded as well as whether the coral had a lesion or the appearance of health. A flow chart of the methods used in this study is provided (Figure 2).
Bacterial Isolation
Samples were plated onto marine agar (MA) and a Vibrio selective media (TCBS)(Difco) plates, as some Vibrio species have been linked to coral disease. Fifty L of seawater, coral mucus, or bag water were pipetted onto each media culture plate and spread using a heat-sterilized glass spreader. The plates were then incubated at room temperature until colonies developed.
Figure 2. A flowchart of project methods.
Single bacterial colonies were collected from the MA and TCBS plates using a variety of selection criteria of visual characteristics such as colony elevation, color, shape, margin, and surface texture. Colonies were selected on the basis of uniqueness relative to other plates, majority of sample, and ease to select single colonies. Selected colonies were transferred into vials containing 1 mL of Zobell's broth (Oppenheimer and Zobell, 1952) and grown overnight at room temperature with gentle agitation to ensure oxygen levels were optimal for growth. After the bacteria were grown for 24 hours, stocks were diluted with a sterile glycerol solution to a final concentration of 60% glycerol and frozen at -80°C for archival purposes. In addition, an aliquot was removed for DNA extractions.
DNA extractions
DNA extraction from individual bacteria colonies was performed using a DNeasy kit (Qiagen, Valencia CA) as per the manufacturers instructions. The protocol for animal tissue was used; cells were chemically lysed and cellular proteins digested using proteinase K and centrifuged onto a microfilter column. Subsequently, DNA was ethanol washed and eluted to a final volume of 100 L using a Tris-buffer provided by Qiagen. The concentration of DNA (g/mL) was determined spectrophotometrically (260 nm).
PCR and Cycle Sequencing
The 16S ribosomal DNA unit was amplified using a Master Taq polymerase PCR kit (Eppendoff, Westbury NY) using 16S-8F primer (5' to 3' AGA GTT TGA TCA TGG CTC AG) and 1513R primer (5' to 3' TAC GGT TAC CTT GTT ACG ACT T) (Sorkin et al., 2001). A 50 µL reaction solution was prepared with 5 µL of 1.5 mM MgCl2 buffer, 1 µL of dNTP's , 0.5 µL of each primer at a concentration of 100 pM/µL , 2.5 U of Taq polymerase, 1 µL of DNA template, and sterile water making up the remaining volume. Samples were run in a thermocycler programmed to cycle 5 minutes at 95ºC prior to 35 repeats of the following steps: 95ºC for 30 seconds, 55ºC for 30 seconds, 72ºC for 1 minute and followed by a final extension step of 72ºC for 3 minutes.
igure 3. Montipora capitata, Porites compressa, and Porites lobata mucus samples divided into seven bacteria categories represented as percentages in the pie charts; excludes Vibrios. N=Number of bacterial sequences used in each species. Symbols: white, actinobacteria; dark gray, alpha; dashed diagonal lines, Cytophaga-flavobacter/flexibacter-bacteroides (CFB); wide gray diagonal lines, firmicutes; light gray, gamma; dashed vertical gray lines, sphingobacteria; medium gray, unknown.
To confirm PCR amplification was successful, the PCR product was run on a 1% agarose gel and DNA was visualized using SYBR green dye (Molecular Probes, Eugene OR) and image capture was performed using a gel-documentation system and software (Quantity One) from BioRad (Hercules, CA).
After the agarose gel confirmed amplification of a PCR product (approximately 350 bp), the amplicon was cleaned up using the PCR purification kit provided by Qiagen (to remove unincorporated dNTPs, polymerase, and PCR reaction components). Recovered DNA was quantitated using a spectrophotometer as previously described.
The PCR amplicon was prepared for sequencing using a Beckman Coulter (Fullerton, CA) protocol: for CEQ Dye terminator cycle sequencing (DTCS) with QuickStart. A 10 µL sequencing reaction was setup using 4 µL of DTCS QuickStart mix, 1 µL of 16S-8F primer (100 pm/µL), and 5 µL of PCR product containing approximately 50 µg DNA. Mineral oil was added on top of the reaction to prevent evaporation during thermocycling. The samples were placed in a thermocycler programmed to cycle at 96ºC for 20 seconds, 50ºC for 20 seconds, and then 60ºC for four minutes and then this cycle was repeated 40 times .
A Millipore Sephadex G-50 (ultrafine) cleanup of the PCR sequencing product was done prior to loading reactions on the Beckman Coulter capillary sequencer.
Data Analysis
The 16S DNA sequences obtained were searched on the National Center for Biotechnology Institution (NCBI) website. The Blast nucleotide-nucleotide search was used to find the first closest match for that sequence (Altschul et al., 1997). Samples that were identified as bacteria were used for ribotype data analysis. Sequences were grouped into taxonomic groups that were based on similar coral disease/bacteria studies (Frias-Lopez et al. 2002, Pantos et al. 2003). The taxonomic groups used were Actinobacteria, Alpha proteobacteria, Gamma proteobacterium, Cytophaga-flavobacter/flexibacter-bacteroides (CFB), Firmicutes, Sphingobacteria, and all others classified as an Unknown category. In part of this study, Vibrio species were excluded from analysis because the two types of media could not be separated out. There would have been a bias towards Vibrios otherwise; thus graphs depicting Vibrio species were left out of the analysis. Based on the dataset of bacterial sequences it was determined that paired samples of M. capitata would provide enough data for the preliminary disease study.
Results
Of the seven bacterial taxonomic groups, four groups (Alpha proteobacteria, Gamma proteobacteria, Actinobacteria, and Unknown bacteria) comprised the bacterial communities found in all three coral species; with the exception of Firmicutes appearing in M. capitata and P. lobata (3%) (Figure 3). Bacterial compositions were distinctly different in all three coral species. For example, surface associated microbes in P. compressa were largely composed of Gamma proteobacteria (67%) and possess a small composition of the Unknown bacteria and Actinobacteria (6%). In contrast to P. compressa, P. lobata had the largest percentage of Actinobacteria (23%). M. capitata was distinguished from the other species due to a larger composition of Unknown bacteria (26%).
All three species were observed to share similarities such as similar representation of Alpha proteobacteria (ranging from 20% to 23%). Also, M. captitata and P. compressa were represented by a low composition of Actinobacteria (6%).
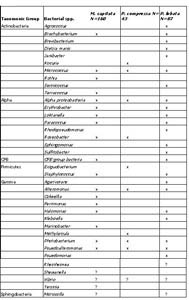
For the breakdown of the taxonomic groups into bacterial species, there were seven bacterial species found in all three corals either as a water or mucus sample (Table 1). The seven bacterial species were Micrococcus, Alpha proteobacteria, Photobacterium, Psuedoalteromonas, Psychrobacter, Vibrios, and Alteromonas. Nine bacterial species were found to occur in at least two of the coral species. All other bacterial species were unique to a specific coral.
Table 1. Unique bacterial species found in Montipora capitata, Porites compressa, Porites lobata bag, mucus and surrounding seawater samples separated into respective taxonomic bacterial groups. N= number of bacterial sequences identified.
An interesting correlation between unique bacterial species and bacterial composition was observed for P. lobata and P. compressa. P. lobata had the highest composition of Actinobacteria in the mucus (23%) (Figure 3) and 5 out of 10 bacterial species (Table 1) from this taxonomic group were unique to water and mucus samples of P. lobata. On the other hand, P. compressa had the highest composition of Gamma proteobacteria (67%) in the mucus and only had one unique bacterial species found in the water and mucus samples.
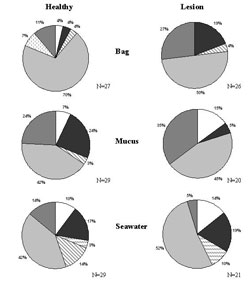
The M. capitata bacterial community in lesioned tissues experienced a shift compared to the healthy M. capitata samples previously discussed. Bacterial shifts observed included an increase in Unknown bacteria and Actinobacteria (Figure 4). Also, there was a decrease in the percentage of Alpha proteobacteria (24% to 5%) found in lesioned tissues.
Initial disease assessment has demonstrated that surface associated microbes of M. capitata are independent of bacterial communities associated with the surrounding seawater. For example, CFB's were only present in the seawater samples, while Firmicutes bacteria were found to be present in higher concentrations in the surrounding water (14%) as compared to the mucus (3%) (Figure 4). Shifts occurred within the Unknown bacteria division where a decrease occurred in the disease samples from 35% in the mucus to 5% in the surrounding seawater samples.
Interestingly, in the bag water there was representation of taxonomic groups not found in the seawater or the mucus of the coral. For example, in the apparently healthy bag water Spingobacteria was represented at 7% and the disease bag water had Firmicutes at 4% that was not seen in mucus or surrounding seawater (Figure 4). Whether this indicates some bacteria are more loosely associated with coral mucus than others requires additional investigation.
Taxonomic breakdown into bacterial species for M. capitata from the lesion samples revealed that water and mucus samples had the same seven bacterial species found previously in all three coral species (Table 2). Four other bacterial species were found in both non-lesion and lesion samples: Erythrobacter, Paracoccus, and CFB group bacteria, and Colwellia. There were 11 out 34 bacterial species unique to the M. capitata lesion samples.
Table 2. Unique bacterial species found in lesioned and non-lesioned Montipora capitata bag, mucus and surrounding seawater samples separated into respective taxonomic bacterial group. N= number of bacterial sequences identified.
Discussion
Bacterial communities associated with the mucus of corals have been shown to be affected by environmental stressors such as increasing sea surface temperatures and coral mucus production (Ritchie and Smith, 1997; Pantos et al., 2003). As corals become stressed due to elevated temperature, there is an increase in mucus production (a coral immune response) and surface associated microbial communities change (Ritchie and Smith 1997). Other factors that may influence the mucus microbial community have yet to be identified. Here we look at how the morphologies may influence the structure of the bacterial communities, based on reports by others that surface structure may influence the establishment of specific bacteria: coral colonization. Speculation by Kim and Ritchie (1997) suggests that Acropora palmata has microbial communities similar to the surrounding water due to its branching morphology and the coral's smaller production of mucus compared to boulder-type corals. The three coral species in our study displayed flatter surface morphologies (P. lobata) and branching morphologies (M. capitata and P. compressa). An overlap of bacterial communities was expected between M. capitata and P. compressa and a distinct different microbial community from P. lobata may be observed due to their inherent differences in physical structure. However, there were no major overlaps of bacterial communities observed between M. capitata and P. compressa. There were nine bacterial species that occurred in both P. lobata and M. capitata; this may be due to the plate morphology each possesses. However, this requires additional investigation, as the samples brought back into the lab were identified as possessing finger versus plate morphology and therefore both structure types may be influencing the microbial communities we observed. Each coral species were distinct from each other, and as Ritchie and Smith (1997) suggest, corals may have microbial communities specific to the species. The only similarities seen were the seven bacterial species that occurred within all three species and the similar composition of the taxonomic group Alpha proteobacteria. This suggests morphology may not influence surface associated microbes and other mechanisms must be in use for corals to obtain their microbial community. To better conclude that morphology isn't influencing bacterial communities, additional research must be accomplished to identify more bacterial sequences for P. compressa and P. lobata to equal out the sample size of M. capitata.
Two species of Vibrio have been identified to cause bleaching in corals (Toren et al., 1998; Rosenberg and Ben-haim, 2002). Toren and collaborators (1998) demonstrated that Oculina patagonica bleached due to a Vibrio strain originally called Vibrio AK-1 (Vibrio shiloi). The bleaching by Vibrio AK-1 affected 80% of the corals in their study after 44 days at an elevated temperature of 26C. In contrast, corals held at a temperature lower than 16C did not bleach. The temperature dependence of Vibrio AK-1 to cause bleaching in corals has also been observed in the other Vibrio (Vibrio corallilyticus) (Kushmaro et al., 2001) that causes bacterial bleaching in Pocillopora damicornis (Rosenberg and Ben-haim, 2002). The study described here is part of a larger study that will attempt to identify which, if any, Vibrio species may be linked to bacterial bleaching in P. damicornis, P. compressa, and P. lobata. Although all Vibrio species identified in this study were grouped into the gamma proteobacteria and not discussed individually, there were more than twenty different types of Vibrio identified by ribotyping. It is beyond the scope of this report to go into more detail about any possible links between specific Vibrio species and bacterial bleaching in Hawaiian corals. However, it is reasonable to expect that if an individual type of Vibrio is isolated routinely from bleached corals, we will be able to apply the methods used by Rosenberg and Ben-Haim (2002) to determine whether Koch' Postulates may be fulfilled.
It is unfortunate that not enough paired samples between each coral type included in this study were collected in order to provide a statistically sound dataset. However, the project is on-going, and as more samples are collected and analyzed, statistics will be performed to determine whether the observations made in this study are truly significant. Another aspect of the study that will be enlarged as more samples are collected will be the types and incidence of lesions occurring in Hawaiian corals throughout the year. The samples analyzed in this current study were all collected in May and early June 2004. Samples will need to be analyzed from collections throughout the year to determine whether some disease occurs more often in the warmer months in Hawaii in which increased irradiance occurs (July-August-September). In the interim, the initial reports in this study are important as a first record of the types of bacteria found associated with coral surfaces in Hawaii.
Figure 4. Montipora capitata non-lesioned and lesioned samples divided into seven bacteria categories represented as percentages in the pie charts; excludes Vibrios. N=Number of bacterial sequences used for each sample type. Symbols: white, actinobacteria; dark gray, alpha; dashed diagonal lines, Cytophaga-flavobacter/flexibacter-bacteroides (CFB); wide gray diagonal lines, firmicutes; light gray, gamma; dashed vertical gray lines, sphingobacteria; medium gray,unknown.
Conclusions
Identifying bacterial associations between Hawaiian corals and their normal and disease-associated microbial communities is an important idea considering coral disease is on an upwards trend globally. This study provides new information of the types of bacteria that are found in the mucus of the three major coral species in the main Hawaiian Islands. Increasing our current knowledge is important because corals are very sensitive to change and they create a habitat for vertebrates and invertebrates to find refuge, mate, and eat. By providing more data to the worldwide data set, conclusions can be drawn about how global warming and anthropogenic impacts are affecting the world's reefs. Increasing the knowledge of how the reefs are currently being impacted may help determine conservation methods that will further sustain important Hawaiian industries that rely on coral ecology such as the fishing industry, recreational, and tourist industries.
Acknowledgements
Support for this work was provided by the Hawaii Coral Reef Initiative and through NSF grant 0243600 and the University of Hawaii Sea Grant College Program. The authors would like to acknowledge the Director of HIMB, Dr. Jo-Ann Leong, for access to all HIMB facilities and David Albert, Lea Hollingsworth, and Dave Krupp for their helpful discussion and technical assistance. Also, the authors would like to acknowledge Sarah Daley of the HIMB core sequencing facility for processing samples. Also, the authors would like to acknowledge our Margaux Kanis for helping with the review/editing process.
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, and Lipman DJ. (1997). Gapped BLAST and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402.
Cooney R, Pantos O, Le Tissier M, Barer M, O'Donnell A, and Bythell J. (2002). Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environmental Microbiology. 4:401-413.
Frias-Lopez J, Zerkle A, Bonheyo G, and Fouke B. (2002). Partitioning of Bacterial communities between Seawater and Healthy, Black Band Diseased, and Dead Coral Surfaces. Applied and Environmental Microbiology. 68:2214-2228.
Harvell CD, Kim K, Burkholder JM, Colwell RR, Esptein PR, Grimes DJ, Hofmann EE, Lipp EK, Osterhaus ADME, Overstreet RM, Porter JW, Smith GW, and Vasta GR. (1999). Emerging Marine Disease,Climate Links and Anthropogenic Factors. Science. 285:16.
Hoover JP. (1998). Hawai'i's Sea Creatures: A guide to Hawai'i's Marine Invertebrates. Honolulu: Mutual.
Kushmaro A, Banin E, Loya Y, Stackebradt E, and Rosenberg E. (2001). Vibrio shiloi sp. nov., the causative agent of bleaching of the coral Oculina patagonica. International Journal of Systematic and Evolutionary Miocrobiology. 51:1383-1388.
Patterson KL, Porter JW, Ritchie KB, Polson SW, Mueller E, Peters EC, Santavy D L, and Smith GW. (2002). The etiology of white pox, a lethal disease of the Caribbean Elkhorn coral Acropora palmate. Ecology. 99:8725-8730.
Oppenheimer CH, and Zobell CE. (1952). The growth and viability of sixty-three species of marine bacteria are influenced by hydrostatic pressure. Journal of Marine Research. 11:10-18.
Sorokin DY, Tourova TP, Lysenko AM, and Kuenen G. (2001). Microbial Thiocyanate Utilization under highly Alkaline conditions. Applied and Environmental Microbiology. 67:528-538.
Sutherland KP, Porter JW, and Torres C. (2004). Disease and immunity in Caribbean and Indo-Pacific zooxanthellate corals. Marine Ecology Progress Series. 266:273-302.
Pantos O, Cooney R, Tissier M, Barer M, O'Donnell A,and Bythell J. 2003. The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastrea annularis. Environmental Microbiology. 370-382.
Ritchie KB, and Smith GW. (1997). Physiological comparison of Bacterial communities from various species of scleractinian corals. Proceeding from 8th International Coral Reef Symposium. 1:521-526.
Rohwer F, Seguritan V, Azam F, and Knowlton N. (2002). Diversity and distribution of coral-associated bacteria. Inter-research. 243:1-10.
Rohwer F, Breithart M, Jara J, Azam F, and Knowlton N. (2001). Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs. 20:85-91.
Smith G. (2004). The Decline of the Coral Reef,Coral Bleaching and Disease with Dr. Garriet W. Smith. Natural Health Museum.
Toren A, Landau L, Kushmaro A, Loya Y, and Rosenberg. (1998). Effect of Temperature on Adhesion of Vibrio strain AK-1 to Oculina patagonica and on Coral Bleaching. Applied and Environmental Microbiology. 64:1379-1384.