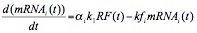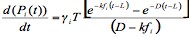Author: Lam Raga Anggara Markely
Institution: Massachusetts Institute of Technology
Date: May 2007
Abstract
Bacteriophage øX174 has been studied for many decades. Nonetheless, no one has ever developed a quantitative model of øX174 development. This model is important to justify the consistency of the literature data and for engineering purposes. Here, a quantitative model is developed for the intracellular øX174 proteins. By using ordinary differential equations, the change in the protein concentration is modeled as the difference between the protein synthesis and degradation rates. The model development uses literature data of the mechanism of øX174 infection process, øX174 genomic sequence, øX174 transcriptional map, øX174 mRNA functional stability, and initial øX174 protein synthesis rate. The model has 4 unknown parameters, including protein degradation rate constant, overall time lag of protein synthesis, a lump parameter that is a product of initial relative concentration of øX174 replicative form DNA, transcription, and translation rate constants, and another lump parameter that is a product of transcription and translation coefficients for cistron A. These parameters are estimated by fitting the model to 42 literature data points of intracellular øX174 protein synthesis rates. Sensitivity analysis is carried out for the protein synthesis rates with respect to øX174 mRNA functional degradation rate constants and øX174 proteins initial translation rates. The results show that the model and experimental data have similar profiles, but there are significant errors between them. The analysis suggests potential sources of errors in the model assumptions and experimental data. The results from this study provide directions for future modeling and experimental studies of the dynamics of øX174 gene expression.
1. Introduction
Bacteriophage øX174 has been studied by scientists in biological areas for many decades. Found in 1920's (Fane et al., 2006), øX174 was the first DNA virus identified to have a single stranded circular genome (Sinsheimer, 1959a, 1959b). In 1961, øX174 DNA, packed in a relatively very small icosahedral molecule, was discovered to be the first infectious viral DNA (Smith et al. 2003). Likewise, øX174 DNA was the first infectious viral DNA to be synthesized with DNA polymerase in 1967 (Goulian and Kornberg, 1967). In 1978, øX174 DNA was sequenced by Frederick Sanger and became the first to be entirely sequenced by Frederick Sanger et al. (1977, 1978). øX174 DNA was found to have 5386 nucleotides and multiple reading frames. Recently, Craig Venter and his colleagues at the Institute for Biological Energy Alternatives created synthetic infectious øX174 DNA and it became the first DNA to be synthesized from scratch (Smith et al. 2003). These and many other studies have provided us with abundant information on the sequence of øX174 DNA, transcriptional map of øX174, the molecular entities involved in the development of øX174, the mechanisms of øX174 development, and the structure of øX174 capsid.
Despite the decades of studies, we still have little quantitative understanding of the intracellular dynamics of øX174 development. Specifically, no one has ever developed a quantitative model of øX174 development that integrates the mechanistic and kinetic data of øX174 development. Without this model, it is not obvious whether the data in the decades of literature are consistent. The quantitative model is also important for engineering purposes, such as phage therapy (Stone, 2002), because we can use it to predict the outcomes of engineering the bacteriophage. Additional examples of the applications of modeling can be found in some of our previous studies: the entry, DNA replication, gene expression, and progeny virus assembly of bacteriophage T7 (Endy et al., 1997), quantitative intracellular dynamics of HIV-1 (Reddy and Yin, 1999), the effects of drugs targeting different viral functions on the growth of bacteriophage T7 (Endy and Yin, 2000), the effects of lipid raft on the dynamics of retroviral entry and trafficking (Lim et al. 2004), and the energetic cost of RNA synthesis and the gene expression of bacteriophage Qβ (Kim and Yin, 2004).
We develop a quantitative model for the intracellular concentration of øX174 proteins. By using ordinary differential equations, we model the change in the protein concentrations as the difference between the protein synthesis and degradation rates. In developing the model, we integrate literature data of the mechanism of øX174 infection process, øX174 genomic sequence, øX174 transcriptional map, øX174 mRNA functional stability, and initial øX174 protein synthesis rate. The unknown parameters in our model are estimated by fitting our model to the experimental data of øX174 protein synthesis rate published by Godson (1971). We also carry out sensitivity analysis, which studies the effect of changing the experimentally measured parameters on the protein synthesis rates. Our analysis suggests several potential sources of errors in the assumptions of our model and the experimental data. In addition, our results can be used as a base for future study in modeling the intracellular dynamics of øX174 gene expression. The analysis of potential sources of error should provide directions in designing experiments for future studies.
The following review is intended to provide a background for our øX174 gene expression model. Since there are limited literature kinetic data, some of the following details are not incorporated into the model. Here, we emphasize on the timing of the process, the viral components involved in each steps of the process, and their corresponding functions. We do not focus on host components since our model assumes that the host resources are unlimited. Readers who are interested in more biological details of øX174 should consult Sinsheimer (1968), Denhardt (1977), Denhardt et al. (1978), Hayashi et al. (1988), and Fane et al. (2006), which cover research results from different periods.
article_1068_order_0
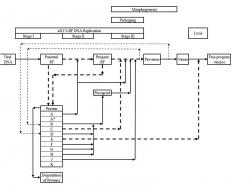
The intracellular øX174 infection process can be divided into 4 major steps, DNA replication, packaging, morphogenesis, and lysis (Fig. 1). These steps take place after the øX174 DNA has been injected into the host cells. øX174 starts the infection by binding to lipopolysaccharide receptors on the E. coli cell surface. Its genome is then ejected from the capsid and enters the host cell (Incardona, 1978). The DNA penetration requires protein H, which is one of the capsid proteins. This protein, also called the DNA pilot protein, penetrates into the cell together with the DNA and directs the binding of the viral DNA (Jazwinski et al. 1975; Jazwinski et al. 1975) to a specific membrane site, at which the viral DNA will attach throughout the infection process (Kozinski, 1961; Sinsheimer et al., 1968). At this site, viral genome undergoes three stages of DNA replication. In stage I DNA replication, viral single stranded (SS) DNA is converted into a double stranded (DS) DNA. This DS DNA is in the form of replicative form (RF) I, which is an RF DNA with both strands closed (Vinograd et al., 1965). The formation of parental RF requires at least 13 host proteins and is totally independent of viral proteins (Hayashi et al. 1988). Stage I DNA replication completes at approximately 3 minutes after infection at 37°C (Sinsheimer, 1968). Once RF I is formed, stage II DNA replication and gene expression take place.
In stage II DNA replication, parental RF replicates through rolling circle mechanism (Eisenberg et al., 1977) to produce progeny RF in the form of RF II, which is an RF DNA with one or more breaks in either one of its strands (Burton and Sinsheimer, 1965). The parental RF serves as the only template for the synthesis of progeny RF and can not be replaced by the newly synthesized progeny RF (Denhardt and Sinheimer, 1965). This process requires the host proteins used in stage I, E. coli rep protein, and viral protein A (Eisenberg et al., 1976). Viral protein A is required to initiate the replication of RF (Baas et al., 1981) by binding and nicking the RF I (Eisenberg et al., 1976; Van Mansfeld et al., 1979) to form RF II-protein A complex (Eisenberg and Kornberg, 1979; Ikeda et al., 1979), terminate the replication process, and regenerate the RF II-rep protein-protein A complex when one round of replication is completed (Brown et al., 1984; Reinberg et al., 1983). At 37°C, the synthesis of progeny RF proceeds at approximately constant rate of 2 molecules/ min until 12-15 minutes after infection, at which approximately 15 progeny RF molecules have been formed (Sinsheimer et al., 1968) and net RF synthesis stops (Sinsheimer, 1968; Sinsheimer et al., 1968). Afterwards, transition from stage II to stage III DNA replication takes place. During this transition, stage II DNA synthesis, host DNA replication and cell division are turned off and stage III DNA synthesis is initiated. This shutdown requires viral protein A* (Eisenberg and Ascarelli, 1981; Funk and Snover, 1976; Langeveld et al., 1979, 1981; Martin and Godson, 1975; Van der Ende et al., 1981, 1982) and protein C, which is discussed below.
Stage III is the production of progeny viral circular SS DNA via rolling circle mechanism. This process is coupled to packaging and morphogenesis (Fig. 1) (Hayashi, 1978), which is described in the following paragraph. At 37°C, the synthesis of progeny SS DNA commences at a rate 5-10 times that of stage II and proceeds at a rate of approximately 1 progeny SS DNA/ (RF. min) (Sinsheimer et al. 1968). This process requires several host proteins, viral protein A, C, J, and procapsid, which consists of viral protein F, G, H, B, and D. The function of viral protein A in stage III is similar to that in stage II. Furthermore, viral protein C is required to inhibit stage II DNA replication (Aoyama and Hayashi, 1986) and associate the procapsid to the template DNA (Aoyama et al. 1981, 1983). This protein specifically binds to one of the DS DNA of an RF II-rep protein-protein A complex with approximately 1:1 stoichiometry. Moreover, this process involves the competition between protein C and host single stranded binding (SSB) protein at the initiation and re-initiation step of stage III DNA replication. If SSB binds first to the RF II-rep protein-protein A complex, stage II DNA synthesis takes place. During this replication, protein C can bind to this replicating complex, but SS DNA will be synthesized only after one round of the replication completes. On the other hand, if protein C binds first, progeny SS DNA will be synthesized provided that procapsid is available (Hayashi et al., 1988). After the progeny SS DNA is packaged into the procapsid, protein C will stay on the RF II-rep protein-protein A-protein C complex to produce more SS DNA (Aoyama and Hayashi, 1986). Viral protein J is required to condense the progeny viral SS DNA in the procapsid (Aoyama et al. 1981, 1983; Hamatake et al. 1985) and prevent stage II DNA replication (Hamatake et al. 1985). This protein enters the procapsid along with the progeny SS DNA during the packaging process (Hamatake et al. 1985). The absence of protein J causes incorrect packaging of the progeny SS DNA (Hamatake et al. 1985), but does not change the synthesis rate of the progeny DNA both in vivo (Hamatake, 1983) and in vitro (Hamatake et al. 1985). Approximately 100 molecules of protein J bind to one molecule of RF DNA template (Hamatake, 1983) and 60 molecules of which will reside in øX174 virion (Burgess, 1969). Protein F, G, and H are major coat protein, major spike protein, and minor spike protein respectively. 12 molecules of protein H, 60 molecules of protein F and G are found in each capsid and procapsid. The other proteins that compose procapsid are viral protein B and D, which are internal and external scaffolding proteins respectively. Each procapsid consists of 60 molecules of protein B and 240 molecules of protein D (Mukai et al. 1979). Both of these proteins are required in morphogenesis.
The first step in morphogenesis is the formation of øX174 provirion, in which protein B is released (Fujisawa and Hayashi, 1977b; Aoyama et al. 1981). øX174 provirion is a molecule that consists of progeny SS DNA, viral protein J, and a procapsid without protein B. Subsequently, the provirion is converted into virion and protein D is dissociated during the process (Fujisawa and Hayashi, 1977a). Finally, progeny viruses are released via lysis, which takes place at approximately 18-25 minutes after infection at 37°C (Sinsheimer, 1968). This process requires the function of viral protein E (Hutchison and Sinsheimer, 1966; Henrich et al. 1982; Young and Young, 1982; Blasi et al. 1985) as well as cell growth (Bradley et al. 1969).
The other viral protein encoded by bacteriophage øX174 is protein K, which is produced at approximately the same amount as protein H (Tessman, et al., 1980). Protein K is a non essential protein for the production of mature phage in vivo, but can affect the burst size of the phage production (Tessman et al. 1980; Gillam, et al. 1985).
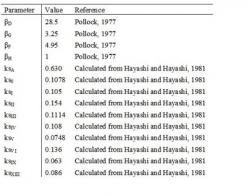
In developing our model, we integrate literature data of the mechanism of øX174 infection process, øX174 genomic sequence (Sanger et al. 1977, 1978), øX174 transcriptional map (Hayashi and Hayashi, 1981), øX174 mRNA functional stability (Fujimura and Hayashi, 1978), and initial øX174 protein synthesis rate (Pollock, 1977). We estimate the unknown parameters in our model by fitting the model to the experimental data of øX174 protein synthesis rates published by Godson (1971). We use the øX174 genomic sequence to calculate the protein synthesis rates from the data reported by Godson (1971). In Godson (1971), labeled amino acids were used to measure the relative protein synthesis rates. The composition of labeled amino acids was, however, not indicated. Given this lack of information, we assume that all labeled 20 amino acids were used in the experiment. In addition, we assume that the reported data represents relative number of labeled amino acids instead of proteins because Godson (1971) does not indicate that the amino acid compositions of each cistron are used in determining the relative protein synthesis rates. Based on the above two assumptions and the composition of amino acids in each cistron from Sanger et al. (1978), we determine the relative protein synthesis rates. Without this calculation of protein synthesis rates, the results of our parameter fitting do not show profiles similar to those of Godson's data (1971) (results not shown). Furthermore, the øX174 transcriptional map is used to determine which mRNA is translated to a certain protein (Appendix). The øX174 mRNA functional stability and initial øX174 protein synthesis rate are used as the values for some of the parameters of our model (Section 2).
In addition to the unknown composition of labeled amino acids, there are other problems in using the data given by Godson (1971). First, Godson (1971) UV-irradiated the host cells prior to infection, which inhibits stage II and III of øX174 DNA replication (Sinsheimer, 1968), while we only have parameters from un-irradiated in vivo experiments. We discuss how to deal with this issue in Section 2. In addition, the cistrons in Godson (1971) are assigned with Roman numerals while the parameters that we obtain from literature correspond to cistrons assigned with alphabets. We translate the Roman numerals to alphabets by comparing the molecular weights, facts regarding the proteins corresponding to the alphabets and Roman numerals, the øX174 genetic map reported in Hayashi et al. (1988) and Sinsheimer (1968). The translation from Roman numerals to alphabets is summarized in Table 1. Despite these problems, we still use Godson's data (1971) because the other data that we find are measured in vitro (Patrushev et al., 1981), show incorrect protein molecular weights and indicate temporal regulation of protein synthesis (Salivar and Loos, 1974), which disagree with other results (Fane, personal communication; Hayashi et al., 1988).
article_1068_order_4
article_1068_order_5
2. Methods and Methods
The øX174 gene expression is modeled with ordinary differential equations. This model is built under the assumption of unlimited host resources. In addition, the UV irradiation of host cells prior to infection is assumed to decrease all protein synthesis rates at different time after infection by the same factor. This assumption allows us to use the parameters from un-irradiated in vivo experiments in parameter fitting with data from irradiated in vivo experiments. The details of the parameter fitting will be discussed in Section 2. The literature parameter values used in the model are summarized in Table 2.
In host cells that are UV irradiated prior to infection, only stage I DNA replication takes place (Sinsheimer, 1968). Given the lack of information regarding RF DNA degradation, the relative concentration of RF is assumed to be constant once the formation of parental RF is completed and can be seen in Equation 1, where t is time in minutes, RF(t) is the relative concentration of RF; RF0 is the initial relative concentration of RF at time t0, which is the time required for stage I of øX174 DNA replication to complete; and Eq. 1 applies for t ≥ t0.
article_1068_order_10
Bacteriophage øX174 exclusively utilizes cis-acting regulation signals that consist of promoters, transcription terminators, mRNA stability sequences, and ribosome binding sites (Fane et al. 2006) to modulate the timing and relative synthesis of its proteins. The first three cis-acting regulation signals are incorporated into the øX174 mRNA model while the ribosome binding sites are incorporated into the øX174 protein model, which is discussed in Section 2. In the dynamics of mRNA, our model accounts the transcription of RF DNA, degradation of mRNA, promoter strength, and transcription termination. In the transcription of DNA, all RF species are assumed to be equally active in transcription. Furthermore, the mRNA is modeled as "bulk mRNA" species corresponding to the cistrons of øX174. These mRNA species are assumed to decay exponentially with half lives that are assumed to be equal to the functional half lives published by Fujimura and Hayashi (1978). In reality, øX174 mRNA exists as mRNAs of different size classes and the structural half lives of these mRNA species have also been determined (Hayashi and Hayashi, 1981). Since the model with size classes of mRNA gives similar results (Appendix) to that with bulk mRNA, we choose the latest as our model for simplicity. The roles of the promoters and transcription terminators on the dynamics of mRNA are assumed to be constant and incorporated into our model as α, which is defined as a transcription coefficient. Based on these assumptions, the rate equation for mRNA can be seen in Equation 2, where subscript i represents cistron D, G, F, A, and H; mRNAi(t) is the relative concentration of mRNA corresponding to cistron i; αi is the transcription coefficient corresponding to cistron i; k1 is the transcription rate constant; and kfi = ln(2)/(t1/2 of mRNAi), is the functional decay constant of mRNAi. Given the relative concentration of RF in Eq. 1 and initial condition of mRNAi(t1) = 0, the solution of Eq. 2 is as follows in Equation 3, which applies for t ≥ t1 ≥ t0,which is the time at which transcription commences.
Equation 2.
article_1068_order_12
The synthesis of øX174 proteins is modeled by accounting the translation of øX174 mRNA, degradation of øX174 proteins, and the ribosome binding sites as the translation modulator. First, we assume that the translation rate constants of each mRNA are equal. We also assume that all proteins have equal degradation rate constants. In addition, we incorporate the translation regulation into our model as β, which is defined as a translation coefficient. With these assumptions, the rate expression of protein synthesis is seen in Equation 4,
article_1068_order_13
where Pi(t) is the relative concentration of protein i; βi is the translation coefficient corresponding to cistron i; k2 is the translation rate constant, and D is the protein degradation rate constant. Given the relative concentration of mRNA in Eq. 3 and the initial condition of Pi(t2) = 0, the first derivative of the solution of Eq. 4 is seen in Equation 5,
article_1068_order_14
where γi ≡ αi.βi; T ≡ k1.k2.RF0; and t2 is the time at which translation commences. To simplify the model, we approximate that transcription and translation start at approximately the same time after infection, i.e. t1 ≈ t2 ≡ L, which is defined as the overall time lag of protein synthesis. This approximation simplifies Eq. 5 to the equation seen in Equation 6,
article_1068_order_15
which applies for t ≥ L. Furthermore, we approximate that γi is proportional to the relative protein synthesis rate at early time after infection determined by Pollock (1977). Given Eq. 6 as our model, we lump the proportionality factor into T and set γi to be equal to these experimentally measured parameters. We also assume that the effects of protein A* and K on the dynamics of øX174 proteins are negligible. In addition, we assume that the amount of protein B and D released from procapsid, if any, is negligible since only the first stage of DNA replication takes place in UV irradiated host cells (Sinsheimer, 1968).
In the parameter fitting, we use the built-in command leasqr.m in GNU Octave to determine the values of the parameters that minimize the difference between the protein synthesis rates obtained from experimental data (Godson, 1971) and predicted by our model. These protein synthesis rates are fitted to Eq. 6 corresponding to five proteins at the same time. Given the lack of kinetic data of protein J, we do not use the data of protein J (Godson, 1971) in the parameter fitting. Hence, our model consists of four fitted parameters, i.e. T, D, L, and γA.
To assess the sensitivity of our øX174 gene expression model to the parameters obtained from literature, we varied γi and t1/2 of mRNAi, one at a time, over a range from 0.01 to 100 times their base case values. With other parameters maintained at their base case values, we run the program to assess the sensitivity of the protein synthesis rate to the modified parameter. In this analysis, we choose the synthesis of protein D as a representation of øX174 gene expression because the model for protein D synthesis rate is similar to that for other proteins, and fits best with the experimental data published by Godson (1971). For this purpose, we define three variables as seen in Equations 7, 8, and 9, where variables with asterisk are those with varied values while the others are maintained at their base case values; Δ indicates the change in protein D synthesis rate due to varying either γD or t1/2 of mRNAD; λ is the ratio of the varied γD to the γD used in the parameter fitting; μ is the ratio of the t1/2 of mRNAD to the t1/2 of mRNAD used in the parameter fitting. This approach allows us to suggest possible causes of the discrepancies between the simulated and experimentally measured results.
3. Results
article_1068_order_16
article_1068_order_1
The profiles of the protein synthesis rates predicted by our model are similar to those in Godson (1971) (Fig. 2). The estimated values of T, D, L, and γA obtained from the parameter fitting are 2.3 min-2, 0.012 min-1, 7.3 min, and 0.74 respectively. In addition, the experimentally measured and predicted protein synthesis rates reach maxima at approximately the same time. There are, however, significant discrepancies between the model and experimental data (Godson, 1971). Among all of the five models, the model for protein D is the closest to the experimental data (Godson, 1971). The synthesis rate of protein D predicted by our model is approximately 0.8 to 2.0 times the experimental values. The synthesis rates of protein G and F predicted by our model are within 0.4 to 1.9 and 1.7 to 3.4 times the experimentally measured data respectively. The model for protein A is within approximately 0.6 to 2.9 times the experimental values at later time. For protein H, the calculated synthesis rate is approximately 2.2 to 16.5 times the reported values (Godson, 1971).
article_1068_order_2
Our sensitivity analysis shows that varying γi increases or decreases protein synthesis rate without changing its profile (Figs. 3A and 3B). Specifically, the protein synthesis rate changes by the same factor that we apply to the translation rate, i.e. Δ = λ for all λ. This behavior is in fact obvious from Eq. 6, which shows that protein synthesis rate is proportional to γi. Furthermore, we find that varying t1/2 of mRNAi not only increases or decreases the protein synthesis rate, but also changes its profile (Figs. 3C and 3D). The protein synthesis rate increases as we increase the functional half life, but unlike in the case of varying γi, the factor by which protein synthesis rate increases is not the same as that applied on functional half life, i.e. Δ ≠ μ. In addition, the peak of the protein synthesis rate is shifted to the right as we increase the functional half life.
We also compare the changes in protein synthesis rate at different time after infection due to variation in t1/2 of mRNAi and γi (Fig. 3E). This comparison summarizes the observations discussed in the previous paragraph. For values of μ close to 1, all the curves and the 45° line coincide. This behavior implies that the change in relative protein synthesis rate due to varying γi follows approximately the same pattern as in the case of varying t1/2 of mRNAi (compare Fig. 3A and 3C). The lines start to diverge at μ approximately equals to 2, indicating that for μ approximately larger than 2, the change in relative protein synthesis rate due to variation in t1/2 of mRNAi is different from that in γi at different times after infection (compare Figs. 3B and 3D). In addition, the change in relative protein synthesis rate due to varying μ decreases in μ (Fig. 3E).
The discrepancies between the model and experimental data indicate that some of the assumptions in the model are inappropriate or there are inconsistencies among the various experimental data that we integrate from literature. The results of our sensitivity analysis suggest that there may be some errors in either our assumption that the labeled amino acids in Godson (1971) consist of all 20 amino acids or the data published by Pollock (1977). Eq.6 indicates that the synthesis rate of protein G should be smaller than that of protein F since γG ≤ γF (Pollock, 1977) and t1/2 of mRNAG ≈ t1/2 of mRNAF (calculated from Fujimura and Hayashi, 1978). However, the opposite is observed in Godson (1971). Our sensitivity analysis shows that increasing γi can raise or lower the protein synthesis rate without changing its profile. Since the profiles of the model and experimental data are similar, errors in the experimental data (Pollock, 1977) that we use as the values of γi can have significant contribution to the observed mismatches. To examine this possibility, we did parameter fitting for protein G and F, one at a time, with γG and γF as the only estimated parameters. In this parameter fitting, we use the previously determined T, D, and L as fixed parameters. We find that the model with new values of γG and γF fit better with the experimental data (Fig. 4). The best fitted γG and γF are 6.26 and 2.66 respectively. Furthermore, if the labeled amino acids do not consist of all the 20 amino acids, our calculation on the relative protein synthesis rates (Section 1) is incorrect and can cause the protein synthesis rate of G to be higher than that of F, instead of the opposite as suggested by Pollock (1977). The above analysis indicates that further justification of our assumption on the labeled amino acids and the data in Pollock (1977) can solve the mismatches between the model and experimental data. The other sources of errors are discussed in Section 4.
4. Discussion
article_1068_order_3
We have developed a mass-action model for the intracellular kinetics of øX174 gene expression. In developing the model, we integrate data from decades of literature: the mechanism of øX174 infection, which are obtained from literature published from 1965 to 1988, øX174 genomic sequence (Sanger et al. 1977, 1978), øX174 transcriptional map (Hayashi and Hayashi, 1981), øX174 mRNA functional stability (Fujimura and Hayashi, 1978), and initial øX174 protein synthesis rate (Pollock, 1977). By fitting our model to the experimental data of øX174 protein synthesis rate (Godson, 1971), we can justify the consistency of the various literature data and the assumptions of our model. Our results show that there are significant discrepancies between the model and experimental data. In the followings, we discuss some of the assumptions of our model and possible experimental errors in Godson's data (1971) that can contribute to the mismatches between the model and experimental data.
We first consider the assumption that UV irradiation prior to infection decreases all protein synthesis rates at different times after infection by the same factor. This assumption is the most fundamental assumption as we try to model a UV irradiated system by using kinetic parameters obtained from un-irradiated systems. This assumption may not be appropriate because in a UV irradiated host, stage II and III DNA replication do not take place (Sinsheimer, 1968). In addition, no SS DNA is produced, which implies that there is no morphogenesis, in which protein B and D are released. Therefore, UV irradiation can affect the dynamics of protein B and D. Given the nature of parameter fitting and that protein D is a major protein, errors in the model of protein D can significantly affect the parameter fitting results of the other proteins. Furthermore, there can also be other unknown changes due to UV irradiation that can affect the dynamics of øX174 protein. Whether these effects are quantitatively significant such that the above assumption should be relaxed is, however, unknown. Further experiments on the effect of UV irradiation on øX174 gene expression should help solve this problem.
Moreover, the continuity assumption across biochemical species, which is inherent to mass-action model, can cause the mismatches. Srivastava et al. (2002) show that this continuity assumption may be misleading for processes that involve low concentration of biochemical species. Since the multiplicity of infection (m.o.i.) of Godson's experiment (1971) is 10, the mismatches between the model and experimental data may be due to this assumption.
We also consider possible errors in the experimental data. As discussed in Section 3, our model (Eq. 6) indicates that, given the data in Pollock (1977), the synthesis rate of protein G should be smaller than that of protein F, but the opposite is observed in Godson (1971). This disagreement on the synthesis rates of proteins F and G is further complicated by the following literature data. Mayol and Sinsheimer (1970) reported that protein F is produced in larger amount than protein G, which is contrary to Godson (1971). On the other hand, the in vitro data published by Patrushev et al. (1981) show that protein G is produced in larger amount than protein F. The results of our sensitivity analysis suggest that further justification of our assumption that the labeled amino acids in Godson (1971) consist of all 20 amino acids and the data in Pollock (1977) can solve the mismatches between the model and experimental data.
It is clear from the above uncertainties in the sources of errors between the model and experimental data that further investigations are required to improve our results. Despite the significant errors, this study has made several steps towards quantitative understanding of the intracellular dynamics of øX174 gene expression. We have integrated mechanistic and kinetic data scattered in decades of literature. In addition, our model can be used as a base for future studies. We also learn several issues that should be considered when designing future experiments for studying the intracellular dynamics of øX174 gene expression: the experiment has to be in vivo because the literature parameter values are obtained from in vivo experiment; the host cells should not be UV-irradiated since some of the stages in øX174 infection can not take place; the multiplicity of infection of the experiment should be sufficiently high so that the error caused by the continuity assumption in the mass-action model can be minimized; and when performing experiments similar to Godson (1971), we must know the composition of the labeled amino acids so that the protein synthesis rates can be calculated accurately.
5. Nomenclature
αi transcription coefficient corresponding to cistron i
βi translation coefficient corresponding to cistron i
γi αi.βi
Δ factor by which protein synthesis rate changes as either γD or t1/2 of mRNAD is varied
λ factor by which γD is varied
μ factor by which t1/2 of mRNAD is varied
D protein degradation rate constant (min-1)
i øX174 cistron index that corresponds to cistron D, G, F, A, and H
k1 transcription rate constant (min-1)
k2 translation rate constant (min-1)
kfi functional decay constant of mRNA corresponding to cistron i (min-1)
L overall time lag of protein synthesis (min)
mRNAi(t) relative concentration of mRNA corresponding to cistron i
Pi(t) relative concentration of protein i
RF(t) relative concentration of øX174 Replicative Form (RF) DNA
RF0 initial relative concentration of øX174 RF DNA
t time (min)
t0 time required for stage I of øX174 DNA replication to complete (min)
t1 time at which transcription starts (min)
t2 time at which translation starts (min)
T k1.k2.RF0 (min-2)
Acknowledgement
The authors would like to thank Bentley A. Fane for providing invaluable information and his review manuscript and Patrick F. Suthers for assisting on computer programming.
Reference
Aoyama, A., Hamatake, R. K., Hayashi, M. (1981) Morphogenesis of øX174: in vitro synthesis of infectious phage from purified viral components. Proc. Natl. Acad. Sci. USA. 78 (12): 7285-7289.
Aoyama, A., Hamatake, R. K., Hayashi, M. (1983) In vitro synthesis of bacteriophage øX174 by purified components. Proc. Natl. Acad. Sci. USA. 80 (14): 4195-4199.
Aoyama, A., Hayashi, M. (1986) Synthesis of bacteriophage øX174 in vitro: Mechanism of switch from DNA replication to DNA packaging. Cell. 47 (1): 99-106.
Baas, P. D., Teertstra, W. R., Van Mansfeld, A. D., Jansz, H. S., Van der Marel, G. A., Veeneman, G. H., Van Boom, J. H. (1981) Construction of viable and lethal mutations in the origin of bacteriophage øX174 using synthetic oligodeoxyribonucleotides. J. Mol. Biol. 152: 615-639.
Blasi, V., Henrich, B., Lubitz, W. (1985) Lysis of Escherichia coli by cloned øX174 gene E depends on its expression. J. Gen. Microbiol. 131 (5): 1107-1114.
Bradley, D. E., Dewar, C. A., Robertson, D. (1969) Structural changes in Escherichia coli infected with a øX174 type bacteriophage. J. Gen. Virol. 5 (1): 113-121.
Brown, D. R., Roth, M. J., Reinberg, D., Hurwitz, J. (1984) Analysis of bacteriophage øX174 gene A protein-mediated termination and reinitiation of øX DNA synthesis. I. Characterization of the termination and reinitiation reactions. J. Biol. Chem. 259 (16): 10545-10555.
Burgess, A. B. (1969) Studies on the proteins of øX174. II. The protein composition of the øX coat. Proc. Natl. Acad. Sci. USA. 64 (2): 613-617.
Burgess, A. B., Denhardt, D. T. (1969) Studies on øX174 proteins. I. Phage-specific proteins synthesized after infection of Escherichia coli. J. Mol. Biol. 44 (3): 377-386.
Burton, A., Sinsheimer, R. L. (1965) The process of infection with bacteriophage øX174. VII. Ultracentrifugal analysis of the replicative form. J. Mol. Biol. 14 (2): 327-347.
Denhardt, D. T. (1977) The isometric single-stranded DNA phages, in: Fraenkel-Conrat, H., Wayner, R. R. (Eds.), Comprehensive Virology, Plenum, New York, pp. 1-104.
Denhardt, D. T. Dressler , D., Ray, D. S. (1978) The single-stranded DNA phages. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York.
Denhardt, D. T., Sinsheimer, R. L. (1965) The process of infection with bacteriophage øX174. V. Inactivation of phage-bacterium complex by decay of 32P incorporated in infecting particle. J. Mol. Biol. 12 (3): 663-673.
Eisenberg, S., Ascarelli, R. (1981) The A* protein of øX174 is an inhibitor of DNA replication. Nucleic Acids Res. 9 (8): 1991-2002.
Eisenberg, S., Griffith, J., Kornberg, A. (1977) øX174 cistron A protein is a multifunctional enzyme in DNA replication. Proc. Natl. Acad. Sci. USA. 74 (8): 3198-
3202.
Eisenberg, S., Kornberg, A. (1979) Purification and characterization of øX174 gene A protein. A multifunctional enzyme of duplex DNA replication. J. Biol. Chem. 254 (12): 5328-5332.
Eisenberg, S., Scott, J. F., Kornberg, A. (1976) An enzyme system for replication of duplex circular DNA: the replicative form of phage øX174. Proc. Natl. Acad. Sci. USA. 73: 1594-1597.
Endy, D., Kong, D., Yin, J. (1997) Intracellular kinetics of a growing virus: A genetically structured simulation for bacteriophage T7. Biotechnol. Bioeng. 55 (2): 375-389.
Endy, D., Yin, J. (2000) Toward Antiviral Strategies that Resist Viral Escape. Antimicrob. Agents Chemother. 44 (4): 1097-1099.
Fane, B. A., Brentlinger, K. L., Burch, A. D., Chen, M., Hafenstein, S., Moore, E., Novak, C. R., Uchiyama, A. (2006) øX174 et al. The Microviridae. In: The Bacteriophages 2nd Edition. pp.129-145. Ed. R. Calendar. Oxford Press.
Freymeyer, D. K. II, Shank, P. R., Edgell, M. H., Hutchison, C. A. III, Vanaman, T. C. (1977) Amino acid sequence of the small core protein from bacteriophage øX174. Biochemistry. 16 (21): 4550-4556.
Fujimura, F. K., Hayashi, M. (1978) Transcription of isometric single-stranded DNA phage, in Denhardt, D. T., Dressler , D., Ray, D. S. (Eds.), The Single-Stranded DNA Phages. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, pp. 485-505.
Fujisawa, H., Hayashi, M. (1977a) Two infectious forms of bacteriophage øX174. J. Virol. 23 (2): 439-442.
Fujisawa, H., Hayashi, M. (1977b) Assembly of bacteriophage øX174: identification of a virion capsid precursor and proposal of a model for the functions of bacteriophage gene products during morphogenesis. J. Virol. 24 (1): 303-313.
Funk, F. D., Snover, D. (1976) Pleiotropic effects of mutants in gene A of bacteriophage øX174. J. Virol. 18 (1): 141-50.
Gillam, S., Atkinson, T., Markham, A., Smith, M. (1985) Gene K of bacteriophage øX174 codes for a protein which affects the burst size of phage production. K. Virol. 53 (2): 708-709.
Godson, G. N. (1971) Characterization and synthesis of øX174 proteins in ultraviolet-irradiated and unirradiated cells. J. Mol. Biol. 57 (3): 541-553.
Goulian, M., Kornberg, A. (1967) Enzymatic synthesis of DNA. 23. Synthesis of circular replicative form of phage øX17 DNA. Proc. Natl. Acad. Sci. USA. 58 (4): 1723-1730.
Hamatake, R. K. (1983) Ph.D. thesis. University of California at San Diego, La Jolla.
Hamatake, R. K., Aoyama, A., Hayashi, M. (1985) The J gene of bacteriophage øX174: In vitro analysis of J protein function. J Virol. 54 (2): 345-350.
Hayashi, M. (1978) Morphogenesis of the isometric phages, in: Denhardt, D. T., Dressler, D., Ray, D. S. (Eds.), The Single-Stranded DNA Phages, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, pp.531-547.
Hayashi, M., Aoyama A., Richardson, D. L., Hayashi, M. N. (1988) Biology of the bacteriophage øX174, in: Calendar, R. (Ed.), Bacteriophages (1st ed.), Plenum, New York, pp. 1-71.
Hayashi, M., Fujimura, F. K., Hayashi, M. (1976) Mapping of in vivo messenger RNAs for bacteriophage øX174. Proc. Natl. Acad. Sci. USA. 73 (10): 3519-3523.
Hayashi, M. N., Hayashi, M. (1981) Stability of bacteriophage øX174-specific mRNA in vivo. J. Virol. 37 (1): 506-510.
Hayashi, M. N., Hayashi M., Imai N. (1981) Bacteriophage øX174 specific messenger RNA synthesis in cells deficient in termination factor ρ activity. J. Virol. 38 (1): 198-207.
Henrich, B., Lubitz, W., Plapp, R. (1982) Lysis of Escherichia coli by induction of cloned øX174 genes. Mol. Gen. Genet. 185 (3): 493-497.
Hutchison, C. A. III, Sinsheimer, R. L. (1966) The process of infection with bacteriophage phi-X174. X. Mutations in a øX Lysis gene. J. Mol. Biol. 18(3): 429-447.
Ikeda, J.-E., Yudelevich, A., Shimamoto, N., Hurwitz, J. (1979) Role of polymeric forms of the bacteriophage øX174 coded gene A protein in øX RFI DNA cleavage. J. Biol. Chem. 254 (19): 9416-9428.
Incardona, N. L. (1978) Adsorption and eclipse reactions of the isometric phages, in: Denhardt, D. T., Dressler, D., and Ray, D. S. (Eds.), The Single-Stranded DNA Phages, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, pp. 549-555.
Iwaya, M., Denhardt, D. T. (1971) The mechanism of replication of øX174 single-stranded DNA. II. The role of viral proteins. J. Mol. Biol. 57 (2): 159-175.
Jazwinski, S. M., Lindberg, A. A., Kornberg, A. (1975) The gene H spike protein of bacteriophages øX174 and S13. I. Functions in phage-receptor recognition and in transfection. Virology 66 (1): 283-293.
Jazwinski, S. M., Marco, R., Kornberg, A. (1975) The gene H spike protein of bacteriophages øX174 and S13. II. Relation to synthesis of the parental replicative form. Virology. 66 (1): 294-305.
Kim, H., Yin, J. (2004) Energy-efficient growth of phage Qβ in Escherichia coli. Biotechnol. Bioeng. 88(2): 148-156.
Kozinski, A. W. (1961) Uniform sensitivity to P32 decay among progeny of P32-free phage øX174 grwon on P32-labeled bacteria. Virology. 13: 377-378.
Langeveld, S. A., Van Mansfeld, A. D. M., De Winter, J. M., Weisbeek, P. J. (1979) Cleavage of single-stranded DNA by the A and A* proteins of bacteriophage øX174. Nucleic Acids Res. 7 (8): 2177-2188.
Langeveld, S. A., Van Mansfeld, A. D. M., Van der Ende, A., Van de Pol, J. H., Van Arkel, G. A., Weisbeek, P. J. (1981) The nuclease specificity of the bacteriophage øX174 A* protein. Nucleic Acids Res. 9 (3): 545-562.
Lim, K. I., Narayan, S., Young, J. A. T., Yin, J. (2004) Effects of lipid rafts on dynamics of retroviral entry and trafficking: Quantitative analysis. Biotechnol. Bioeng. 86 (6): 650-660.
Martin, D. F., Godson, G. N. (1975) Identification of a bacteriophage øX174 coded protein involved in the shut-off of host DNA replication. Biochem. Biophys. Res. Commun. 65 (1): 323-330.
Mayol, R. F., Sinsheimer, R. L. (1970) Process of infection with bacteriophage øX174. XXXVI. Measurement of virus-specific proteins during a normal cycle of infection. J. Virol. 6 (3): 310-319.
Mukai, R., Hamatake, R. K., Hayashi, M. (1979) Isolation and identification of bacteriophage øX174 prohead. Proc. Natl. Acad. Sci. USA. 76 (10): 4877-4881.
Patrushev, L. I., Kapitza, E. L., Stukacheva, E. A., Shemyakin, M. F. (1981) Control of bacteriophage øX174 gene expression by the transcription termination factor ρ in vitro. Mol. Gen. Genet. 182 (3): 471-476.
Pollock, T. J. (1977) Gene D of bacteriophage øX174: Absence of transcriptional and translational regulatory properties. J. Virol. 21 (2): 468-474.
Pollock, T. J., Tessman, E. S., Tessman, I. (1978) Identification of lysis protein E of bacteriophage øX174. J. Virol. 28 (1): 408-410.
Reddy, B., Yin, J. (1999) Quantitative intracellular kinetics of HIV-1. AIDS Res. Hum. Retroviruses. 15 (3): 273-283.
Reinberg, D., Zipursky, S. L., Weisbeek, P., Brown, D., Hurwitz, J. (1983) Studies on the øX174 gene A protein-mediated termination of leading strand DNA synthesis. J. Biol. Chem. 258 (1): 529-537.
Salivar, W. O., Loos, L. J. (1974) Evidence for the temporal regulation of protein synthesis in synchronized bacteriophage øX174 infections. J. Gen. Virol. 23 (3): 309-330.
Sanger, F., Air, G. M., Barrell, B. G., Brown, N. L., Coulson, A. R., Fiddes, J. C., Hutchison, C. A. III, Slocombe, P. M., Smith, M. (1977) Nucleotide sequence of bacteriophage øX174. Nature. 265 (5596): 687-695.
Sanger, F., Coulson, A. R., Friedmann, T., Air, G. M., Barrell, B. G., Brown, N. L., Fiddes, J. C., Hutchison, C. A. III, Slocombe, P. M., Smith, M. (1978) The nucleotide sequence of bacteriophage øX174. J. Mol. Biol. 125 (2): 225-246.
Sinsheimer, R. L. (1959a) Purification and properties of bacteriophage øX174. J. Mol. Biol. 1 (1): 37-42.
Sinsheimer, R. L. (1959b) A single-stranded deoxyribonucleic acid from bacteriophage øX174. J. Mol. Biol. 1 (1): 43-53.
Sinsheimer, R. L. (1968) Bacteriophage øX174 and related viruses. Prog. Nucleic Acid Res. Mol. Biol. 8: 115-169.
Sinsheimer, R. L., Knippers, R., Komano, T. (1968) Stages in the replication of bacteriophage øX174 DNA in vivo. Cold Spring Harb. Symp. Quant. Biol. 33: 443-447.
Smith, H. O., Hutchison, C. A. III, Pfannkoch, C., Venter, J. C. (2003) Generating a synthetic genome by whole genome assembly: øX174 bacteriophage from synthetic oligonucleotides. Proc. Natl. Acad. Sci. USA. 100 (26): 15440-15445.
Srivastava, R., You, L., Summers, J., Yin, J. (2002) Stochastic versus deterministic modeling of intracellular viral kinetics. J. Theor. Biol. 218: 309-321.
Stone, R. (2002). Stalin's forgotten cure. Science 298: 728-731.
Tessman, E. S, Tessman, I., Pollock, T. J. (1980) Gene K of bacteriophage øX174 codes for a nonessential protein. J. Virol. 33 (1): 557-560.
Van der Ende, A., Langeveld, S. A., Teertstra, R., Van Arkel, G. A., Weisbeek, P. J. (1981) Enzymatic properties of the bacteriophage øX174 A protein on superhelical øX174 DNA: A model for the termination of the rolling circle DNA replication, Nucleic Acids Res. 9 (9): 2037-2053.
Van der Ende, A., Langeveld, S. A., Van Arkel, G. A., Weisbeek, P. J. (1982) The interaction of the A and A* proteins of bacteriophage øX174 with single-stranded and double-stranded øX DNA in vitro. Eur. J. Biochem. 124 (2): 245-252.
Van Mansfeld, A. D., Langeveld, S. A., Weisbeek , P. J., Baas, P. D., Van Arkel, G. A., Jansz, H. S. (1979) Cleavage site of øX174 gene-A protein in øX and G4 RFI DNA. Cold Spring Harb. Symp. Quant. Biol. 43: 331-334.
Vinograd, J., Lebowitz, J., Radloff, R., Watson, R., Laipis, P. (1965) The twisted circular form of polyoma viral DNA. Proc. Natl. Acad. Sci. USA. 53 (5): 1104-1111.
Young, K. D., Young, R. (1982) Lytic action of cloned øX174 gene E. J. Virol. 44 (3): 993-1002.
Appendix
This model is similar to Eq. 6 except that the mRNA is modeled as size classes of mRNA that decays exponentially with structural half lives obtained from Hayashi and Hayashi (1981). There are in fact two sources (Fujimura and Hayashi, 1978; Hayashi and Hayashi, 1981) that publish two different sets of structural decay constants, but we use the more recent data (Hayashi and Hayashi, 1981) in our model since both papers have the same co-author. In order to determine the mRNA species that correspond to each protein, we use the øX174 transcription map (Hayashi and Hayashi, 1981), which is summarized in Table A.1. With these assumptions, the relative protein synthesis rate is given in Equation A.1, where j is the index of protein; i is the index of the mRNA species; Pj(t) is the relative concentration of protein j; βj is the translation coefficient corresponding to cistron i; ksi is the structural decay constant corresponding to mRNA species i; T, D, t, and L are the same as those of Eq. 6. The kinetics parameters used in this model are summarized in Table A.2.
article_1068_order_7
article_1068_order_8
The results of the parameter fitting by using this model (Fig. A.1) are similar to those discussed in Section 3. The estimated T, D, L, and βA are 0.22 min-2, 0.016 min-1, 7.1 min, and 9.6 respectively. Although the model for protein H (Eq. A.1) fits better with the experimental data (Figs. 2 and 6), we decide to use the model given by Eq. 6 because the improvement is insignificant and Eq. 6 is simpler than Eq. A.1.
article_1068_order_9