Authors: Michael Workman, Olivia D. Nigro, and Grieg F. Steward
Institution: Ohio University
Date: November 2006
ABSTRACT
Staphylococcus aureus is an opportunistic human pathogen that is found in a large percentage of the population and passively colonizes primarily in the skin or in the nares. It is also occasionally found in high abundance in coastal recreational waters due to shedding from swimmers and other beachgoers. As a result, coastal swimming areas are a potential source of community-acquired S. aureus infections. The capacity of a given S. aureus strain to cause infection varies among strains and is determined by the presence or absence of a large number of virulence-associated genes, some of which are encoded by prophages. In order to determine the abundance and characterize the diversity of S. aureus strains to which swimmers in coastal waters of Oahu may be exposed, 30 isolates were collected from seawater at Kuhio Beach in Waikiki, HI and screened for the presence or absence of phage-type specific DNA segments using polymerase chain reaction. One to five prophage segments were detected per isolate with no prophage-free strain found. Prophage segments from ø3A-like and ø77-like phages were most common, showing up in nearly all isolates collected. Data from this study suggests that prophage content in S. aureusisolates varies widely among strains and the diversity at Kuhio Beach is relatively high. The results provide insight into the role of bacteriophages in the changing ecology and virulence of S. aureus.
INTRODUCTION
Staphylococcus aureus is a Gram-positive bacterium and an opportunistic human pathogen that has been a major topic of study for the past several decades due to the increasing emergence of new virulent and antibiotic resistant strains throughout hospital and community settings (Chambers 2001). Symptoms of a S. aureus infection depend in part on the route of infection and on the array of virulence-associated genes that are present and expressed by the infecting strain. Manifestations of infection may include scalded skin syndrome, deep abscesses, gastroenteritis, and potentially lethal toxic shock syndrome (Boyd and Brüssow 2002). Several sources indicate that approximately 20-30% of the general population is infected, primarily on the skin or in the nasal passages (Centers for Disease Control and Prevention 2005; Kluytmans and Wertheim 2005; Werthiem et al. 2005). S. aureus is also present in recreational coastal waters as a result of swimmers who readily shed the bacterium from their nose, skin, and respiratory track (Charoenca and Fujioka 1993). Data from Charoenca and Fujioka (1993) shows that S. aureus is present in coastal waters around Oahu, Hawaii at mean concentrations between < 2 CFU/ml and 92.9 CFU/ml which are significantly correlated with swimmer density. Their results imply that marine waters serve as a potential route for the spread of S. aureus and that exposure to staph-contaminated water may result in colonization or infection.
The mechanism by which many new and pathogenic strains of S. aureus have emerged is believed to be mainly due to horizontal gene transfer of virulence genes; often by plasmids, transposons, insertions sequences, pathogenicity islands, and bacteriophages (Boyd et al. 2002; Wilson and Salyers 2003). Boyd and colleagues (2002) conclude that bacteriophages play the leading role in the horizontal transfer of virulence genes and have the ability to convert non-pathogenic strains of bacteria into virulent strains through phage lysogenic conversion. In this process, the transfer and incorporation of bacteriophage DNA into the bacterial host as a prophage can result in increased ecological fitness for the bacterium through the gained ability to colonize new host tissues, evade host immune responses, and produce new toxins. The presence of prophages in the genome of a given strain of S. aureuscan be detected by amplification of phage-specific gene fragments using the polymerase chain reaction (PCR). Many of the S. aureus-associated bacteriophages contribute to the ill effects of infection by introducing new genes into the bacterium which alter its capacity to produce various toxins and virulence factors such as enterotoxins A, E, G, K, and P, staphylokinase, β-lysin and lipase, exfoliative toxin A, panton-valentine leukocidin, and toxic shock syndrome toxin-1 (Boyd and Brüssow 2002; Boyd et al. 2001; Pantucek et al. 2004).
Peacock et al. (2002) note the common behavior of S. aureus to act either as a harmless commensal or a serious pathogen as a result of the variable presence of disease-causing virulence genes within a given population of S. aureus. This suggests that discrimination among closely related S. aureus strains would be useful when evaluating water quality since total abundance may not directly correlate with risk. To our knowledge, very few studies have been published characterizing the genetic composition of S. aureus isolates collected from tropical coastal waters. A recent preliminary study found that isolates of S. aureus from coastal recreational waters of Oahu had variable combinations of virulence-associated genes (Stotts et al. 2004). In this complementary study, the spatial variability and abundance of S. aureus was determined at a single beach on Oahu, Hawaii and thirty new isolates were characterized for their prophage content.
METHODS AND MATERIALS
Collection of S. aureus
Surface ( < 6 cm depth) seawater samples from Kuhio Beach in Waikiki, Hawaii were collected in triplicate on May 30, 2005 and June 28, 2005 at various distances from shore, on both the seaward and shoreward sides of a seawall that runs parallel to the beach and encloses a recreational swimming area roughly 50m X 200m. On both sampling occasions, samples were collected from an onshore-offshore transect originating near the center of the beach and consisting of four collection sites: the nearshore site was within one meter of shore in water ≤ 0.1 m deep, the second site was midway between the shoreline and the seawall in water having a depth of about 1 m, the third site was directly adjacent to the seawall on the shoreward side with a depth of approximately 1.5 m, and the fourth site was adjacent to the wall on the seaward side. For the June sampling, two additional nearshore sites were sampled at the far right and far left ends of the beach to test for alongshore variability. Samples were collected by pumping 100 ml of seawater through silicone tubing into Nalgene® bottles using a hand operated water pump. Prior to each collection, the tubing was sterilized with ethanol then rinsed with 3 volumes of sample water to remove any residual ethanol remaining in the tubing. Samples were placed in a cooler with ice packs and transported to the lab where they were filtered through 0.2 µm PVDF membrane filters (Millipore Corporation, Billerica, Massachusetts). Filters were placed face up on CHROMagar S. aureus selective medium (CHROMagar Microbiology, Paris, France) and allowed to incubate at 37ºC for 24 hours. Putative S. aureus colonies were picked and transferred to a second CHROMagar plate to verify that their color was mauve/purple, which is indicative of S. aureus on CHROMagar, as opposed to blue, yellow, green, cream, or colorless (Galliot et al. 2000). The mauve/purple colonies were then serially streaked onto fresh CHROMagar or trypticase soy agar plates using an inoculating loop to ensure isolation of clonal populations for archiving and DNA extraction.
Isolation of S. aureus DNA
Isolated S. aureus colonies were inoculated into 1.5 ml of trypticase soy broth (TSB) and incubated at 37ºC for 18 hours under constant agitation. After incubation, the contents were centrifuged at 10,000 x g for 10 min followed by removal of the supernatant and resuspension of the pellet in 600 µl of TER buffer containing 0.5 g of 0.1 mm zirconium beads (Biospec Products, Inc. Bartlesville, Oklahoma). The mixture was placed in a FastPrep bead beater (Q-Biogene, Carlsbad, California) for two 30s cycles at a setting of 6 m/s. 75 µl of SDS (10%) and 120 µl of 5M NH4OAc was added to the mixture and then incubated on ice for 30 min. The contents were centrifuged at 14,000 x g for 10 min before transferring the supernatant into 800 µl of isopropanol and incubating on ice for an additional 30 min to help precipitate the DNA. The mixture was then centrifuged at 10,000 x g for 15 min at 4ºC, the isopropanol was aspirated off, and the remaining pellet was washed with 70% ethanol. The contents were centrifuged for a final cycle at 10,000 x g for 15 min at 4ºC, the ethanol was aspirated off, and the samples were allowed to air dry before resuspension in 100 µl of TE buffer.
Quantification of Isolated S. aureus DNA
DNA concentrations were determined using a TD-700 filter-based laboratory fluorometer (Turner Designs Inc., Sunnyvale, California) and a Quant-iTTM DNA assay quantification kit (Molecular Probes Inc., Eugene, Oregon) following the protocols provided with each. After quantification, DNA stocks were diluted to a 10 ng µl-1 working concentration using TE buffer.
Individual Primer Pair PCR
article_844_order_2
A 25 µl reaction mixture was made consisting of 50 ng of template DNA, PCR buffer (1X), 200 µM dNTP mixture, 2 mM MgCl2, 1.5 units Platinum Taq Polymerase, one of the following primers pairs: SGB1/SGB2, SGD1/SGD2, SGL1/SGL2, SGFa1/SGFa2, SGFb1/SGFb2, or SGA1/SGA2 (Table 1), and enough nuclease free water to reach the 25 µl volume. The primer pairs were designed from conserved genomic sequences from phage types ø3A, ø11, ø77, ø187, and Twort (Pantucek et al. 2004). The primers for Twort-like phages served as a negative control since the Twort phage is a lytic phage in the family Myoviridae and does not integrate into the host chromosome. Each mixture was placed in a thermal cycler (iCycler, BioRad, Hercules, CA) for initial denaturation (5 min, 94ºC) and 30 cycles of amplification consisting of denaturation (1 min, 94ºC), annealing (1 min, 58ºC), and chain extension (1 min, 70ºC). Amplification was followed by one final chain extension cycle (3 min, 70ºC) and electrophoresis of 8 μl of PCR product and 2 μl of 5X loading buffer in a 2.0% agarose gel for 1 hour at 90 V in 0.5X TBE buffer.
Multiplex PCR
A 25 μl reaction volume was mixed following the protocol set by Pantucek et al. (2004), which consisted of 50 ng of template DNA, PCR buffer (1X), 200 μM dNTP mixture, primers SGB1/SGB2, SGD1/SGD2, SGL1/SGL2, and SGFb1/SGFb2 (0.3µM each), SGA1/SGA2 (0.6 µM each), and SGFa1/SGFa2 (0.8 µM each), 2 mM MgCl2, 1.5 units Platinum TaqPolymerase, and enough nuclease-free water to reach the 25 μl volume. Primers SAU1 and SAU2 (0.3 µM each), derived from a 44-kb conserved region present in the genome of S. aureus, were also added to the multiplex reactions to serve as a positive control. Each reaction mixture was then loaded into a thermal cycler for initial denaturation (5 min, 94ºC) and 30 cycles of amplification consisting of denaturation (1 min, 94ºC), annealing (1.5 min, 58ºC), and chain extension (1.5 min, 70ºC). Amplification was once again followed by a final chain extension cycle (3 min, 70ºC) and electrophoresis of 8 μl of PCR product and 2 μl of 5X loading buffer in a 2.0% agarose gel for 1 hour at 90 V in 0.5X TBE buffer.
RESULTS
Abundance of S. aureus in coastal water
article_844_order_1
The site average concentrations of putative S. aureus CFU (colony forming units) ranged from 1 to 14 CFU per 100 ml in May and from 8 to 177 CFU per 100 ml in June (Figure 1). The highest count for a single sample was 448 CFU per 100 ml for one of the nearshore replicates collected in June. Besides this one outlier all other samples had concentrations of ≤ 58 CFU per 100 ml. On both sampling occasions, the number of putative S. aureus CFU was highest in the shoreline samples and decreased with increasing distance from the shore. The number of CFU per 100 ml was significantly higher in the June sampling at all sites excluding the site directly inshore of the seawall (p < 0.05).
Identification of Prophages in Coastal Water S. aureus Isolates
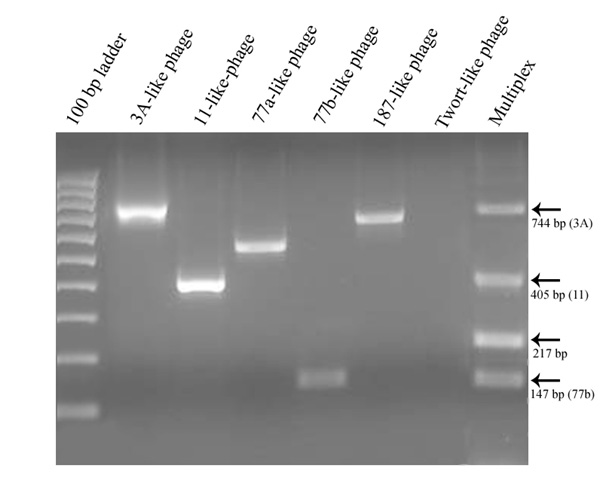
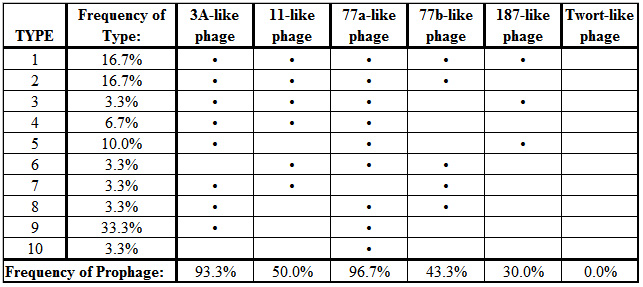
All of the targeted prophage types, with exception of the Twort-like phage, were successfully amplified by the polymerase chain reaction when primer pairs were used individually. However, some of the PCR products failed to amplify reliably when the primer sets were combined into a multiplex reaction (Figure 2). Of the 30 isolates that were characterized, a total of 10 different combinations of prophage carriage were found, ranging from one to five prophage types detected per isolate (Table 2). The most commonly detected prophages belonged to the ø77a-like and ø3A-like phage groups with carriage rates of 96.7% and 93.3% respectively.
article_844_order_0
article_844_order_3
DISCUSSION
Abundance of S. aureus in coastal waters
In a previous study of staphylococci in coastal waters, Charoenca and Fujioka (1993) classified Kuhio Beach as a "high staph" site with S. aureus concentrations ranging from < 2 to 472 CFU per 100 ml (mean of 77.5 CFU per 100 ml). This is very similar to range of concentrations observed in this study, though the overall mean calculated for all samples was higher in their study, probably because we included offshore sites in this study. The average for our nearshore sites (72 CFU per 100 ml) is very similar to that reported by Charoenca and Fujioka (1993). Since viable S. aureus have been consistently found on every sampling occasion these waters appear to be a persistent source of exposure for swimmers. Charoenca and Fujioka (1993) observed that the concentration of S. aureus in marine water is significantly linked to swimmer density and similar findings were seen in this study. S. aureus concentrations were higher at the time of the June sampling than the May sampling as was the number of swimmers (85 in May vs. 125 in June). Although S. aureus concentrations correlate with swimmer density, it is possible that other variables such as wave height and flushing rate within the swimming area may have contributed to the higher counts in June. It was observed during the June sampling, for example, that fewer waves were spilling over the seawall than in May, which may have resulted in a lower flushing rate. The occasional very high concentrations found in this and the previous study illustrates that concentrations are both spatially and temporally variable, and exposure risk may be higher than indicated by average values.
Identification of Prophages using Multiplex PCR
Multiplex PCR can offer a rapid and convenient method of identifying multiple gene targets in a single reaction and, in this study, a multiplex method for detection of the known major prophage groups of S. aureus (Pantucek et al. 2004) was attempted. Unfortunately, the reliability of the multiplex assay could not be established for the isolates in this study. Most of the isolates characterized by multiplex PCR were missing amplification products relative to the results from individual primer pairs. The ø187-like target, in particular, rarely amplified in multiplex reactions, but was readily detected when assayed individually. Lowering of the annealing temperature resulted in non-specific products with no improvement in the number or type of prophages that could be detected. However, other adjustments to the multiplex assay such as the use of a hot-start Platinum Taq DNA polymerase (Invitrogen Life Technologies Corporation, Carlsbad, California) and increased MgCl2 concentration from 1 mM to 2 mM did improve the sensitivity of the multiplex reaction by allowing detection of up to four prophages in one isolate, but the results never corresponded consistently with those obtained with single primer pair reactions. Routine assay of the isolates was therefore performed using independent PCR reactions for each targeted prophage.
Identification of Prophages in Coastal Water S. aureus Isolates
As was found in this study, Pantucek et al. (2004) observed that every S. aureus isolate assayed harbored at least one prophage segment and many had more than one. The prophage content per bacterium, however, tended to be higher in this study of coastal seawater isolates than was found in the 176 clinical isolates assayed by Pantucek et al. (2004). For example, 16.7% of S. aureus isolates from Kuhio Beach was positive for all five prophage groups and 20% were positive for four. In contrast, Pantucek et al. (2004) did not observe any isolates containing all five prophage types, and only about 1% contained four types. The most frequently detected prophage groups (ø3A-like and ø77a-like) were the same in this study and that of Pantucek et al. (2004), but the specific combinations of prophage types present within individual isolates varied substantially in both studies. The high frequency of prophage carriage and diversity in prophage content among strains of S. aureus support the idea that phages play a major role in the changing ecology and virulence of this pathogen.
Broader Implications
Differences in the virulence gene content of S. aureus have been correlated with the manifestations of disease (Peacock et al. 2002) and bacteriophages are one of the major mechanisms mediating lateral transfer of virulence-associated genes in S. aureus and other pathogens. The documentation of high diversity in S. aureus isolates from coastal waters, both by direct detection of virulence-associated genes (Stotts et al. 2004) and by detection of prophages (this study), suggests that recreational coastal waters are sites at which humans are exposed to many new strains of S. aureus of varying capacity to colonize or cause disease. Characterization of the coastal reservoir of S. aureus strains and their associated phages is an important first step in assessing the role of coastal seawater in the transmission and ecology of this rapidly evolving pathogen. Future research to determine the probability of colonization or infection as a result of contact with staph-contaminated water would be a useful next step in developing water quality guidelines for heavily used, tropical coastal waters.
ACKNOWLEDGEMENTS
Support for the MSURF program was provided by the National Science Foundation (Grant #02-43600), the University of Hawaii Sea Grant College Program, and the Pacific Research Center for Marine Biomedicine at the University of Hawaii. Research support was provided by grants from NSF (OCE04-32479) and NIEHS (1P50EF012740-01). The author gratefully acknowledges the mentoring of Dr. G.F. Steward, O.D. Nigro, and Dr. M.J. Cooney.
REFERENCES
Baba, T. et al. (2002). Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 359: 1819-1827.
Boyd, E.F. and H. Brüssow (2002). Common themes among bacteriophage-encoded virulence factors and the diversity among bacteriophages involved. TRENDS in Microbiology.10: 521-529.
Centers for Disease Control and Prevention (2005). Community-Associated MRSA information for clinicians. [http://www.cdc.gov/ncidod/dhqp/ar_mrsa_ca_clinicians.html] link current as of May 2006.
Chambers, H.F. (2001). The changing epidemiology of Staphylococcus aureus? Emerging Infectious Diseases. 7: 178-182.
Charoenca, N. and R.S. Fujioka (1993). Assessment of Staphylococcus bacteria in Hawaii's marine recreational waters. Water Science and Technology. 27: 283-289.
Coleman, D.C. et al. (1989). Staphylococcus aureus bacteriophages mediating the simultaneous lysogenic conversion of β-lysin, staphylokinase and enterotoxin A: molecular mechanism of triple conversion. J Gen Microbiology. 135: 1679-1697.
Galliot, O. et al. (2000). Evaluation of CHROMagar Staph. aureus, a new chromogenic medium, for isolation and presumptive identification of Staphylococcus aureus from human clinical specimens. Journal of Clinical Microbiology. 38: 1587-1591.
Henegariu, O. et al. (1997). Multiplex PCR: Critical parameters and step-by-step protocol. Biotechniques. 23: 504-511.
Kluytmans, J.A.J.W. and H.F.L. Wertheim (2005). Nasal carriage of Staphylococcus aureusand prevention of nosocomial infections. Infection. 33: 3-8.
Kuroda, M. et al. (2001). Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 357: 1225-1240.
Pantucek, R. et al. (2004). Identification of bacteriophage types and their carriage in Staphylococcus aureus. Archives of Virology. 149: 1689-1703.
Peacock, S.J. et al. (2002). Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infection and Immunity. 70: 4987-4996.
Robinson, D.A. and M.C. Enright (2004). Multilocus sequence typing and the evolution of methicillin-resistant Staphylococcus aureus. Clinical Microbiology and Infectious Diseases.10: 92-97.
Stotts, S. N. et al. (2005). Virulence and antibiotic resistance gene combinations among Staphylococcus aureus isolates from coastal waters of Oahu, Hawaii. The Journal of Young Investigators 12: 4. [http://www.jyi.org/research/re.php?id=148] link current as of May 2006.
Wertheim, H.F.L. et. al (2005). The role of nasal carriage in Staphylococcus aureus infections. The Lancet Infectious Diseases. 5: 751-762.
Wilson, B.A. and A.A. Salyers (2003). Is the evolution of bacterial pathogens an out of body experience? TRENDS in Microbiology. 11: 347-350.