Authors: Joseph Granata, Hugo Sanchez, Phillip Loeschinger, and Jodi F. Evans
Institution: Molloy College, 1000 Hempstead Ave, Rockville Centre, NY 11570
ABSTRACT
Mesenchymal progenitor cells are widely studied for their ability to regulate macrophage cell responses and can be suppressive or supportive during an inflammatory response. Mouse aorta-derived progenitors (mAo) support the macrophage inflammatory response and highly express CD105, also known as endoglin. Elevated CD105 expression is consistently associated with inflammatory disease. Therefore, we hypothesized that suppression of CD105 will reduce the immunosupportive capacity of mAo mesenchymal progenitors. We used siRNA to reduce expression of CD105 in mAo cells. We subsequently examined the effect of this deficiency in their response to both lipopolysaccharide (LPS) and their ability to support the macrophage inflammatory response. Contrary to our hypothesis, after exposure to LPS, CD105-deficient mAo secreted increased levels of the inflammatory indicators nitric oxide (NO) and interleukin 6 (IL- 6). This occurred with both mAo that were cultured alone and in co-culture with macrophage. The observed increase of NO and IL-6 in the co-cultures is additive and therefore points to the mAo as the primary origin. Overall, our data suggest that CD105 acts as a regulator of the TLR-4 pathway and may represent an important target for modification of mesenchymal progenitors to be used in therapeutics.
INTRODUCTION
The development and use of cell-based therapeutics is at the forefront of modern medicine, and mesenchymal progenitor cells are a major focus of investigation. Mesenchymal progenitor cells are the multipotent precursors to connective tissue cells (Young et al., 1995) and have traditionally been studied due to their role in tissue repair after damage. They are also capable of regulating immune cell function through direct and indirect cell contact making them a novel tool in the treatment of many inflammatory diseases (Aggarwal & Pittenger, 2005). They can modulate the activity of many immune cell types including macrophages and can be immunosupportive or immunosuppressive. Some mesenchymal progenitors influence macrophages by alternating their orientation from the inflammatory M1 to the anti-inflammatory M2 phenotype, rendering these progenitors immunosuppressive (Cho et al., 2014; Fernandez et al., 2017). Other studies show that mesenchymal progenitors are pro-inflammatory when co-cultured with macrophages making these progenitors immunosupportive (Anton, Banerjee, & Glod, 2012; Evans, Salvador, George, Trevino-Gutierrez, & Nunez, 2015). The phenotypic characteristics associated with these properties are an active area of study and their expression of toll-like receptors (TLR) are thought to play a role. TLR transmembrane protein receptors are sensors of microorganisms and have a critical function in innate immunity (Waterman, Tomchuck, Henkle, & Betancourt, 2010).
Many studies have also focused on the role of surface antigens. The surface antigens CD73, CD90 and CD105 are used to identify mesenchymal progenitors (Dominici et al., 2006), but also are likely to participate in their regulation of immune cells. Previous research groups have reported that mesenchymal progenitors derived from mouse aorta (mAo) highly express CD90 and CD105, but do not express CD73 (Fernandez et al., 2017). This population of mesenchymal progenitors (mAo), when in co-culture with macrophages, promotes inflammation (Evans et al., 2015). In contrast, a mouse bone marrow derived cell line lacking expression of CD90 and CD105 suppresses macrophage activity (Fernandez et al., 2017). The mechanisms mediating this opposing regulation are yet to be determined but because CD105 is highly expressed in chronically inflamed tissues (Middleton et al., 2005), our goal was to focus on its role in the mAo’s ability to support the macrophage inflammatory response.
CD105 or endoglin is a co-receptor for the TGFβ superfamily of receptors. TGFβ receptors are known for their function in regulating cell growth and differentiation of cells (Lin & Moustakas, 1994). Due to alternative splicing of the CD105 transcript, there is both a long isoform (L-CD105) and short isoform (S-CD105) of the CD105 protein (Dallas et al., 2008). The two isoforms share identical sequences (Bellon et al., 1993; Gougos & Letarte, 1990); however, L-CD105 contains an extra sequence leading to functional differences. Functionally, L-CD105 decreases type I TGFβ receptor-induced collagen production and increases type I TGFβ receptor-induced cell proliferation while the short isoform performs the opposite function. It greatly enhances type I TGFβ receptor-induced collagen and decreases cell proliferation (Velasco et al., 2008).
In this study, we hypothesized that if expression of CD105 is reduced in mAo cells, their support of the macrophage inflammatory response would be reduced. The ability of CD105-deficient mAo to respond to the toll-like receptor – 4 (TLR-4) ligand, LPS, and regulate the inflammatory response of MΦ was therefore examined. Defining a role for CD105 in mesenchymal progenitor regulation of macrophage activity will aid in the identification and isolation of specific progenitor subsets for use in targeted cell-based therapeutics.
MATERIALS AND METHODS
Materials
All cell culture media, trypsin, FBS and antibiotic/antimycotic solutions were obtained from Invitrogen (Carlsbad, CA). The PE rat anti-mouse CD105 (Cat. # 562759) was purchased from BD Biosciences. Gamma irradiated LPS from E. coli (#L4391) and all other chemicals and reagents were purchased from SIGMA-Aldrich (USA) unless otherwise specified.
Animals
All animal protocols were approved by the Winthrop University- Hospital’s Animal Care and Use Committee and adhere to the regulations outlined by the National Institutes of Health. C57BL/6 male mice were obtained from Taconic, North America. Animals were housed under local vivarium conditions (12 h light-dark cycle) and allowed to acclimate for at least 7 days prior to experimentation. Mice were euthanized under CO2 at 8-12 weeks of age and aorta and hind limbs were removed in preparation for cell isolation.
Cell Culture
Aortic mesenchymal progenitor cell line (mAo)
Mouse aortic progenitor lines were derived from C57BL/6 mice using the method of da Silva Mierelles (da Silva Meirelles, Chagastelles, & Nardi, 2006) with slight modification as described in (Evans, Fernando, & Ragolia, 2012).
Bone marrow-derived macrophage cells (MΦ)
Bone marrow from the hind limbs of the C57BL/6 mouse was isolated as previously described (Yeh, Evans, Chen, & Aloia, 1999). After creating a single-cell suspension, nucleated cells were counted using 3% acetic acid/trypan blue exclusion and plated in DMEM supplemented with 10% fetal bovine serum (FBS), 15% L929 fibroblast cell conditioned medium, 100 U/mL penicillin sodium, 100 U/mL streptomycin sulfate and 0.25 μg/mL amphotericin B at 107 cells per 100 mm petri dish. The L929 conditioned medium was prepared as suggested by the American Type Culture Collection (ATCC). The L-929 cell line produces M-CSF which supports the growth and differentiation of macrophages from the bone marrow. After three days, half the medium was removed and replaced with fresh medium. A complete medium change was performed on day 6. At day 7 of culture, monocyte/macrophage cells were re-plated according to experimental objectives, passed or frozen and stored in liquid nitrogen (LN2). Cultures up to passage two were used in experiments.
Co-culture
The mAo cells were initiated at a density of 1.5 x 10^4 cells/cm^2. On the fifth day of post transfection with CD105 siRNA, MSCs cultures reached confluence, and MΦ were added at a density of 1.0 x 10^5 cells/cm^2 and allowed to attach overnight. Cultures were then left untreated or were treated with LPS (100 ng/ml) for 24 h. Culture supernatants were then collected and stored at -80ºC until assay.
siRNA Transfection
Transfection was carried out using Silencer® Select siRNA (siRNA ID # s65509) directed to the endoglin (CD105) gene (Ambion, Life Technologies) with the sequence 5’-GGACGGUGACCGUAAUGAUtt-3’. Lipofectamine RNAiMAX (Invitrogen) was used to transfect the mAo cells according to the manufacturer’s suggestions.
Immunoblotting (Western Blot)
Cells were lysed using RIPA buffer (Santa Cruz Biotechnology, Inc.) and protein lysates were then quantified using the Bradford assay (Pierce). Protein lysates, 50 μg per well, were subjected to SDS-PAGE using a premade (4-20%) polyacrylamide gel (Invitrogen). Separated proteins were transferred onto a PVDF membrane. Membranes were blocked with 5% nonfat milk in TBST, and then primary endoglin antibody (Novus/Gene ID 2022/Host rabbit) was diluted in 5% BSA/TBST and incubated overnight. The secondary antibody, HRP-conjugated anti-rabbit IgG (Santa Cruz Biotechnology, Inc.), was then applied and detection was carried out using enhanced chemiluminescence (ECL) detection reagent (ThermoFisher Scientific). Expression was normalized using β-Actin.
Nitrite Measurements
Nitrite, a reflection of NO production, was measured in cell culture supernatant using the Griess Reagent system (Promega, Madison, WI) according to the manufacturer’s instructions.
Real-time reverse transcription PCR
Real-time reverse transcription PCR was carried out using Sso Advanced Universal SYBR Green supermix and the CFX96 Touch Real-Time PCR System from Bio-Rad. Primer sequences were specific for mouse as used by (Perez-Gomez et al., 2005); L-Eng forward 5’-CGACTCTGGTACATCTATTCTCACACACGTGG-3’ L-Eng reverse 5’–GGGCACTACGCCATGCTGCTGGTGG-3’, S-Eng forward 5’–TGAGTATCCCAAGCCTCCACCCCAT-3’ S-Eng reverse 5’–CTGAGGGGCGTGGTGAAGGTCAG-3’. PCR conditions were 95ºC for 5 minutes followed by 40 cycles of 95ºC 10s, 55ºC 10 s, and 72ºC for 30 s.
Statistical Analyses
Unless indicated otherwise, data were analyzed using one-way ANOVA. Post hoc test P values were adjusted by using the Bonferroni correction. All tests were two-tailed, and a nominal significance level of 0.05 was used.
RESULTS
Relative expression of short and long isoforms of CD105 in mAo progenitor cells
Figure 1. Relative expression of L-CD105 and S-CD105 in mAo pro-genitors. A) Gene expression was measured using real-time reverse transcription PCR and expression normalized to the GAPDH gene. The inset shows the raw amplification curve. B) Protein expression was measured using Western blot and quantified through densitometry. The inset shows a representative blot. Results are presented as mean ± SD with n = 4.
First, the relative expression of S-CD105 and L-CD105 in the mAo progenitor cells was established at both the transcript and protein levels, using real-time reverse transcription PCR and western blotting, respectively (Figure 1). This was done to determine if the mAo express both isoforms of CD105 and to establish baseline expression. In mAo progenitor populations, the basal expression of L-CD105 is ~2.6-fold greater than S-CD105 at the transcript level (Figure 1A) and ~2.33-fold greater at the protein level (Figure 1B).
Optimization of siRNA protocol
Figure 2. Optimizing the siRNA transfection protocol. A) mAo progenitors were transfected with a range of CD105 siRNA concentrations (5-30 nmole) and transfection efficiency on days 2, 4, and 5 post-transfection was confirmed by Western blot. B-C) Densitometry was used to quantify expression of the long (B) and short (C) forms of CD105 at the protein level.
The siRNA transfection protocol was subsequently optimized to determine the optimal concentration of siRNA as well as the timeline for post-transfection incubation. A range of siRNA concentrations were evaluated (5 - 30 nM) along several post-incubation time points, days 2, 4, and 5. At a 30 nM concentration of siRNA and five days post-transfection, both S-CD105 and L-CD105 were reduced to ~29% and 40% of control, respectively (Figure 2). The results from immunoblotting were confirmed via quantitative means using real-time RT-PCR. RNA was extracted from cultures that were exposed to a 50nm concentration of siRNA and remained exposed for a 5-day post-transfection incubation period. The real-time RT-PCR results indicated that the expression of L-CD105 was reduced 4.81-fold and the expression of S-CD105 was increased 1.91-fold after siRNA exposure (Figure 3).
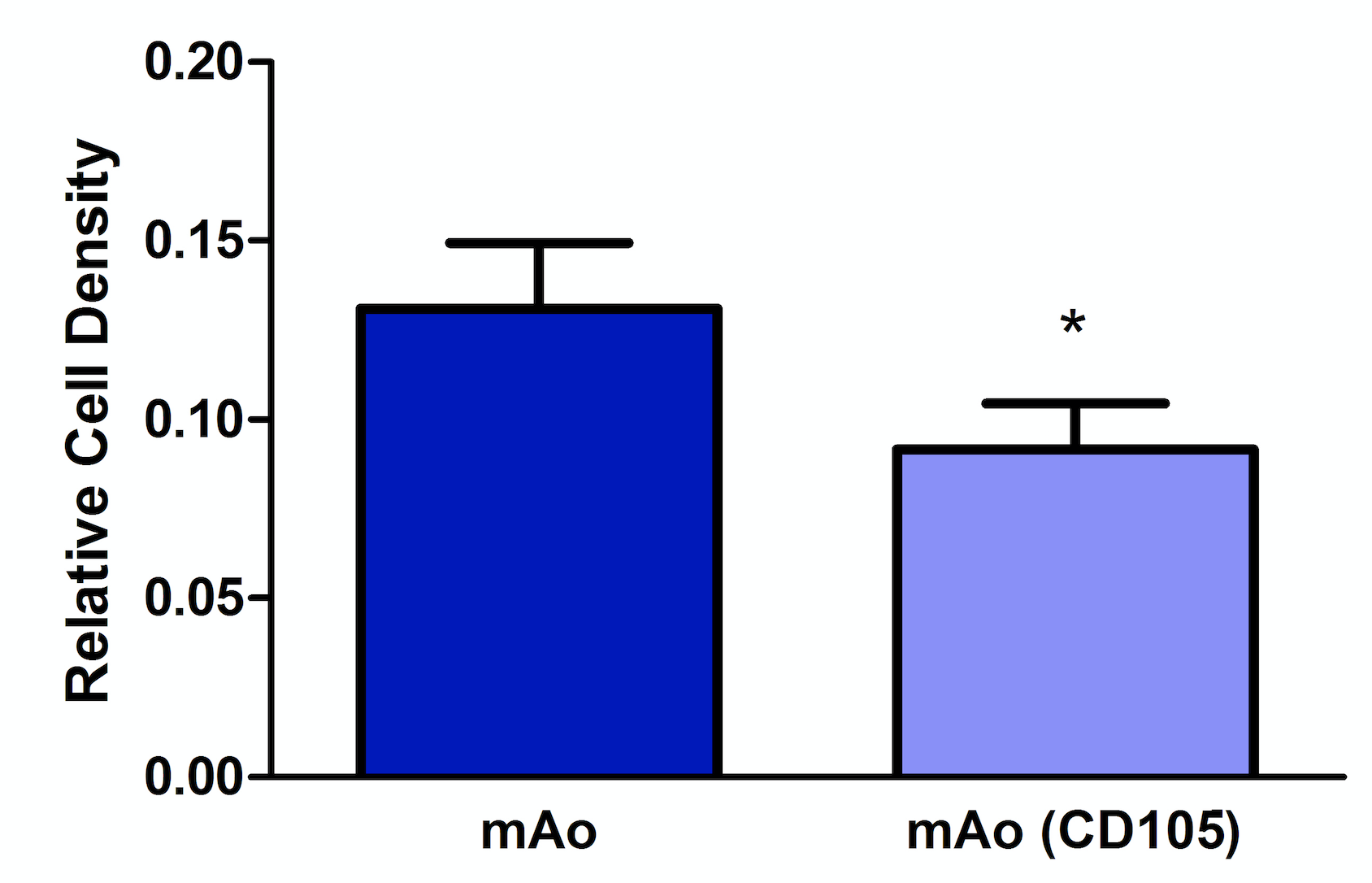
Reduction of CD105 expression results in decerased mAo cell density
CD105 expression is associated with cellular proliferation; therefore, we examined the effect of CD105 deficiency on cell density of mAo. These data were necessary to distinguish between density related effects or mechanistic related effects of CD105 deficiency in mAo during an inflammatory response. CD105-deficient mAo cultures had a significantly lower cell density, ~1.4-fold, when compared to the control cultures (Figure 4). These data suggest that CD105 deficiency results in a decreased ability of mAo to proliferate.
CD105-deficient mAo significantly increase expression of NOand IL-6 in response to LPS
Figure 3. Confirmation of siRNA transfection efficiency. Using real-time reverse transcription PCR and CD105 isoform-specific primers, transcript levels of S-CD105 and L-CD105 were measured in mAo and mAo transfected with CD105 siRNA. Data are expressed as mean ± SEM and are representative of three separate experiments.
mAo progenitors synergistically enhance the response of MΦ to the inflammatory mediator, LPS (Evans et al., 2015). Therefore, the next step was to determine how CD105 deficiency affects the inflammatory response of mAo and how it affects the way they regulate the inflammatory response of MΦ. CD105-deficient mAo were cultured alone and co-cultured with MΦ followed by exposure to LPS. NO and IL-6 production were used as indicators of inflammation. When in co-culture with MΦ, mAo synergistically enhance NO and IL-6 production in response to LPS (Figure 5). CD105-deficient mAo respond to LPS with an increased production of NO and IL-6 when compared to control mAo cultures, p < 0.001 and p < 0.05, respectively. Similarly, CD105-deficient mAo/ MΦ co-cultures produce increased levels of NO and IL-6 compared to control mAo/MΦ co-cultures, p < 0.001 and p < 0.05, respectively. This increase in NO and IL-6 is additive and is contributed by the CD105-deficient mAo progenitors.
Figure 4. Reduction of CD105 expression results in a decreased mAo progenitor cell density. Cell density was measured on day 5 post-transfection using methylene blue. Data are expressed as mean ± SEM, and are representative of three separate experiments each with n = 6. *Statistical significance as determined by t-test.
CD105 deficiency does not significantly alter TLR-4 gene expression in mAo cells
When exposed to LPS, CD105-deficient mAo respond with significantly increased expression of inflammatory mediators, p < 0.05. To determine if an upregulation of the LPS receptor, TLR-4, is responsible for this increased inflammatory response we used real-time reverse transcription PCR. While TLR-4 is expressed in mAo cells, depletion of CD105 does not significantly alter its transcription (Figure 6).
Figure 5. LPS-induced NO (A) and IL-6 (B) production is increased in supernatant of CD105-deficient mAo progenitor cultures and in co-cultures of CD105-deficient mAo and MΦ. Data are expressed as mean ± SEM and are representative of three separate experiments each with n = 6. Significant differences were determined after 2-way ANOVA using Bonferroni’s post-test.
DISCUSSION
Figure 6. CD105 deficiency does not alter TLR-4 expression in mAo. Gene expression of TLR-4 was measured in mAo progenitors treated with and without 50 nm siRNA. Data are expressed as mean ± SEM and are representative of three separate experiments.
Contrary to our hypothesis, the reduction of CD105 expression did not reduce the inflammatory response of the mAo cell after exposure to LPS nor did it result in a decrease of NO and IL-6 in when mAo were in co-culture with MΦ. In fact, reduced expression of CD105 in mAo cells resulted in increased production of these inflammatory markers. This effect appears mechanistic rather than related to changes in cell density as cell density was reduced in mAo cultures treated with CD105 siRNA. This reduction in proliferation is consistent with studies in other cell types where CD105 expression is associated with supporting proliferation. In endothelial cells, CD105 promotes TGFβ/ALK1 signaling and inhibits TGFβ/ALK5 signaling thus promoting endothelial cell proliferation and differentiation (Waterman et al., 2010). Additionally, CD105 enriched periodontal ligament mesenchymal stem cells (PDLSMSCs) exhibit an increased proliferative capacity (Albiero et al., 2015). However, there is no difference in proliferation capacity among CD105 negative and CD105 positive subsets of mouse adipose-derived progenitors (Anderson, Carrillo-Galvez, Garcia-Perez, Cobo, & Martin, 2013). These data taken together point to CD105 having different roles in progenitor cell proliferation dependent on cell origin.
The short and long isoforms of CD105 have been shown to yield opposing effects on cell proliferation; the short isoform inhibits cell proliferation while the long isoform enhances cell proliferation (Velasco et al., 2008). It was observed in our research that mAo progenitor cells express both the short and long isoforms of CD105 with the long isoform predominating. These data show the deficiency of the long isoform of CD105 as the mechanism through which mAo cell proliferation is decreased and is consistent with previous data that demonstrate CD105 expression and cell proliferation are directly correlated (Lebrin et al., 2004).
When confirming the siRNA-induced decreased protein expression of the short and long isoforms of CD105 at the transcript level, it was anticipated that the results would parallel one another, but this was not the case. The decrease in L-CD105 transcript mirrored the reduction in protein and was reduced 4.81-fold. However, the decrease in S-CD105 protein was not reflected in the gene expression studies, and there was a 1.91-fold increase in the S-CD105 transcript. One potential cause for the discrepancy between protein and transcript levels of S-CD105 could be related to the complex process of protein synthesis. Eukaryotic transcription and translation is not only complex, but the two processes occur in different locations within the cell (Berg, Tymoczko, & Stryer, 2002). Both the complexity of these processes and the movement between organelles may have led to the difference in measured protein and transcript levels of S-CD105. Another reason for the discrepancy could be because these two isoforms differ from one another in the composition of their cytoplasmic tails (Bellon et al., 1993; Gougos & Letarte, 1990; Perez-Gomez et al., 2005). This means that the sequence of S-CD105 is contained within the sequence of L-CD105. Upon knocking down L-CD105, the remaining sequence could have been that of S-CD105, resulting in an apparent increase of S-CD105.
NO and IL-6 are secreted by mAo and MΦ in response to LPS and in mAo/MΦ co-cultures there is a synergistic increase in NO and IL-6 production in response to LPS (Evans et al., 2015; Fernandez et al., 2017). Decreased CD105 expression led to an even greater secretion of these inflammatory markers in both mAo cultured alone and mAo in co-culture with MΦ. This increase was an additive increase contributed by the CD105-deficient mAo. These data are in contrast to previous studies which examined the interaction of mouse adipose-derived CD105 negative progenitors and macrophage cells. In these studies, both the CD105 positive and CD105 negative adipose derived cells promoted the regulatory macrophage phenotype and reduced the inflammatory profile (Anderson et al., 2013). One potential explanation is that inflammatory marker production in response to LPS could vary depending on the source of the progenitor cell. The progenitors in this study were derived from mouse aorta while the progenitors of the contrasting study were derived from adipose tissue. Future studies using both aorta and adipose tissue are required to confirm these differences.
Like other progenitors derived from the vasculature (Guijarro-Munoz, Compte, Alvarez-Cienfuegos, Alvarez-Vallina, & Sanz, 2014), mAo highly express TLR-4. The fact that CD105-deficient mAo respond to LPS with increased inflammatory marker production suggests that CD105 acts as a regulator of the TLR-4 pathway in these cells. However, transcription of TLR-4 was not changed in CD105-deficient mAo. Although we did not confirm that CD105 deficiency resulted in changes in protein levels of TLR-4 in these studies, they point to CD105 regulation of signaling molecules downstream of TLR-4. Additional studies to determine how CD105 interacts with the TLR-4 pathway are warranted. The identification of mechanisms through which CD105 regulates this pathway would contribute to the development of new anti-inflammatory and immunosupportive cell-based therapeutics.
CONCLUSION
mAo progenitor cells express both the short and long isoform of CD105 with the long isoform predominating. Expression of CD105 in these cells plays a role in both their proliferation and inflammatory response and points to CD105 regulation of the TLR-4 pathway. Therefore, CD105 may represent an important target for modification of mesenchymal progenitors to be used in therapeutics.
ACKNOWLEDGMENTS
This work was supported by Molloy College Biology, Chemistry, and Environmental Studies (BCES) Department and by a grant from the NIH/NHLBI, R00HL091116-05 to JFE.
REFERENCES
Aggarwal, S., & Pittenger, M. F. (2005). Human mesenchymal stem cells modulate allogeneic immune cell responses. Blood, 105(4), 1815-1822. doi:10.1182/blood-2004-04-1559
Albiero, M. L., Amorim, B. R., Martins, L., Casati, M. Z., Sallum, E. A., Nociti, F. H., Jr., … Martin, F. (2013). CD105 (endoglin)-negative murine mesenchymal stromal cells define a new multipotent subpopulation with distinct differentiation and immunomodulatory capacities. PLoS One, 8(10), e76979. doi:10.1371/journal.pone.0076979
Anton, K., Banerjee, D., & Glod, J. (2012). Macrophage-associated mesenchymal stem cells assume an activated, migratory, pro-inflammatory phenotype with increased IL-6 and CXCL10 secretion. PLoS One, 7(4), e35036. doi:10.1371/journal.pone.0035036
Bellon, T., Corbi, A., Lastres, P., Cales, C., Cebrian, M., Vera, S., … Bernabeu, C. (1993). Identification and expression of two forms of the human transforming growth factor-beta-binding protein endoglin with distinct cytoplasmic regions. Eur J Immunol, 23(9), 2340-2345. doi:10.1002/eji.1830230943
Berg, J. M., Tymoczko, J. L., & Stryer, L. (2002). Eukaryotic Transcription and Translation are Separated in Space and Time. Biochemistry (5th edition ed.). New York: W.H. Freeman.
Cho, D. I., Kim, M. R., Jeong, H. Y., Jeong, H. C., Jeong, M. H., Yoon, S. H., . . . Ahn, Y. (2014). Mesenchymal stem cells reciprocally regulate the M1/M2 balance in mouse bone marrow-derived macrophages. Exp Mol Med, 46, e70. doi:10.1038/emm.2013.135
da Silva Meirelles, L., Chagastelles, P. C., & Nardi, N. B. (2006). Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci, 119(Pt 11), 2204-2213. doi:10.1242/jcs.02932
Dallas, N. A., Samuel, S., Xia, L., Fan, F., Gray, M. J., Lim, S. J., & Ellis, L. M. (2008). Endoglin (CD105): a marker of tumor vasculature and potential target for therapy. Clin Cancer Res, 14(7), 1931-1937. doi:10.1158/1078-0432.CCR-07-4478
Dominici, M., Le Blanc, K., Mueller, I., Slaper-Cortenbach, I., Marini, F., Krause, D., …Horwitz, E. (2006). Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy, 8(4), 315-317. doi:10.1080/14653240600855905
Evans, J. F., Fernando, A., & Ragolia, L. (2012). Functional melanocortin-2 receptors are expressed by mouse aorta-derived mesenchymal progenitor cells. Mol Cell Endocrinol, 355(1), 60-70. doi:10.1016/j.mce.2012.01.019
Evans, J. F., Salvador, V., George, S., Trevino-Gutierrez, C., & Nunez, C. (2015). Mouse aorta-derived mesenchymal progenitor cells contribute to and enhance the immune response of macrophage cells under inflammatory conditions. Stem Cell Res Ther, 6, 56. doi:10.1186/s13287-015-0071-8
Fernandez, N., Renna, H., McHugh, L., Mazolkova, K., Crugnola, W., & Evans, J. F. (2017). Mouse Mesenchymal Progenitor Cells Expressing Adipogenic and Osteogenic Transcription Factors Suppress the Macrophage Inflammatory Response. Stem Cells Int, 2017, 5846257. doi:10.1155/2017/5846257
Gougos, A., & Letarte, M. (1990). Primary structure of endoglin, an RGD-containing glycoprotein of human endothelial cells. J Biol Chem, 265(15), 8361-8364.
Guijarro-Munoz, I., Compte, M., Alvarez-Cienfuegos, A., Alvarez-Vallina, L., & Sanz, L. (2014). Lipopolysaccharide activates Toll-like receptor 4 (TLR4)-mediated NF-kappaB signaling pathway and pro-inflammatory response in human pericytes. J Biol Chem, 289(4), 2457-2468. doi:10.1074/jbc.M113.521161
Lebrin, F., Goumans, M. J., Jonker, L., Carvalho, R. L., Valdimarsdottir, G., Thorikay, M., . . .Girard, J. P. (2005). A comparative study of endothelial cell markers expressed in chronically inflamed human tissues: MECA-79, Duffy antigen receptor for chemokines, von Willebrand factor, CD31, CD34, CD105 and CD146. J Pathol, 206(3), 260-268. doi:10.1002/path.1788
Perez-Gomez, E., Eleno, N., Lopez-Novoa, J. M., Ramirez, J. R., Velasco, B., Le tarte, M., . . . Quintanilla, M. (2005). Characterization of murine S-endoglin isoform and its effects on tumor development. Oncogene, 24(27), 4450-4461. doi:10.1038/sj.onc.1208644
Velasco, S., Alvarez-Munoz, P., Pericacho, M., Dijke, P. T., Bernabeu, C., Lopez-Novoa, J. M., & Rodriguez-Barbero, A. (2008). L- and S-endoglin differentially modulate TGFbeta1 signaling mediated by ALK1 and ALK5 in L6E9 myoblasts. J Cell Sci, 121(Pt 6), 913-919. doi:10.1242/jcs.023283
Waterman, R. S., Tomchuck, S. L., Henkle, S. L., & Betancourt, A. M. (2010). A new mesenchymal stem cell (MSC) paradigm: polarization into a pro-inflammatory MSC1 or an Immunosuppressive MSC2 phenotype. PLoS One, 5(4), e10088. doi:10.1371/journal.pone.0010088
Yeh, J. K., Evans, J. F., Chen, M. M., & Aloia, J. F. (1999). Effect of hypophysectomy on the proliferation and differentiation of rat bone marrow stromal cells. Am J Physiol, 276(1 Pt 1), E34-42.
Young, H. E., Mancini, M. L., Wright, R. P., Smith, J. C., Black, A. C., Jr., Reagan, C. R., & Lucas, P. R. (1995). Mesenchymal stem cells reside within the connective tissues of many organs. Dev. Dyn., 202, 137-144. doi:10.1002/aja.1002020205